PUBLICATIONS
2023

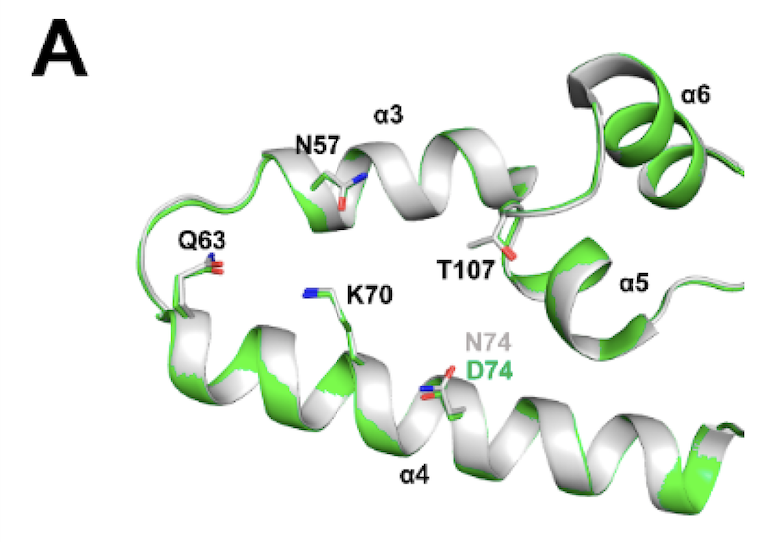
175. Multidisciplinary studies with mutated HIV-1 capsid proteins reveal structural mechanisms of lattice stabilization. 197. Gres AT, Kirby KA, McFadden WM, Du H, Liu D, Xu C, Bryer AJ, Perilla JR, Shi J, Aiken C, Fu X, Zhang P, Francis AC, Melikyan GB, Sarafianos SG. Nat Commun. 2023 Sep 12;14(1):5614. DOI:10.1038/s41467-023-41197-7. PMID: 37699872
Localization of NS5A and dsRNA following treatment with CsA and LDV.

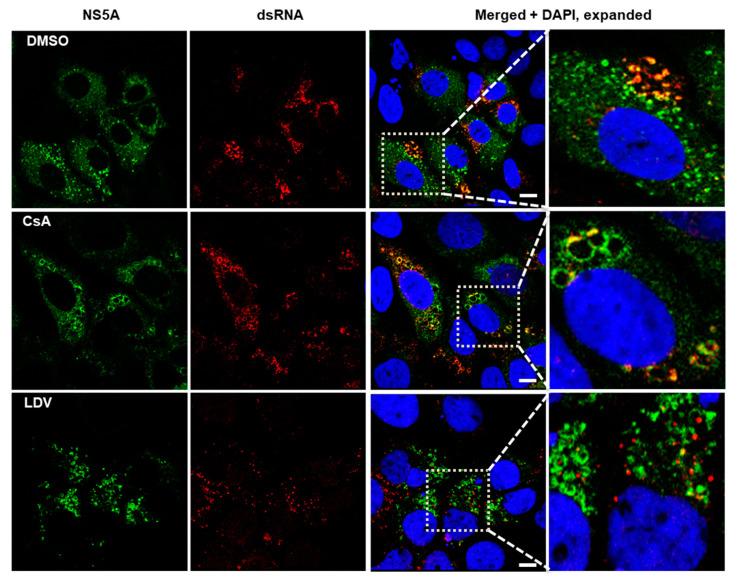
174. Mechanisms of Action of the Host-Targeting Agent Cyclosporin A and Direct-Acting Antiviral Agents against Hepatitis C Virus. Liu D, Ndongwe TP, Ji J, Huber AD, Michailidis E, Rice CM, Ralston R, Tedbury PR, Sarafianos SG. Viruses. 2023 Apr 17;15(4):981. doi: 10.3390/v15040981. PMID: 37112961.
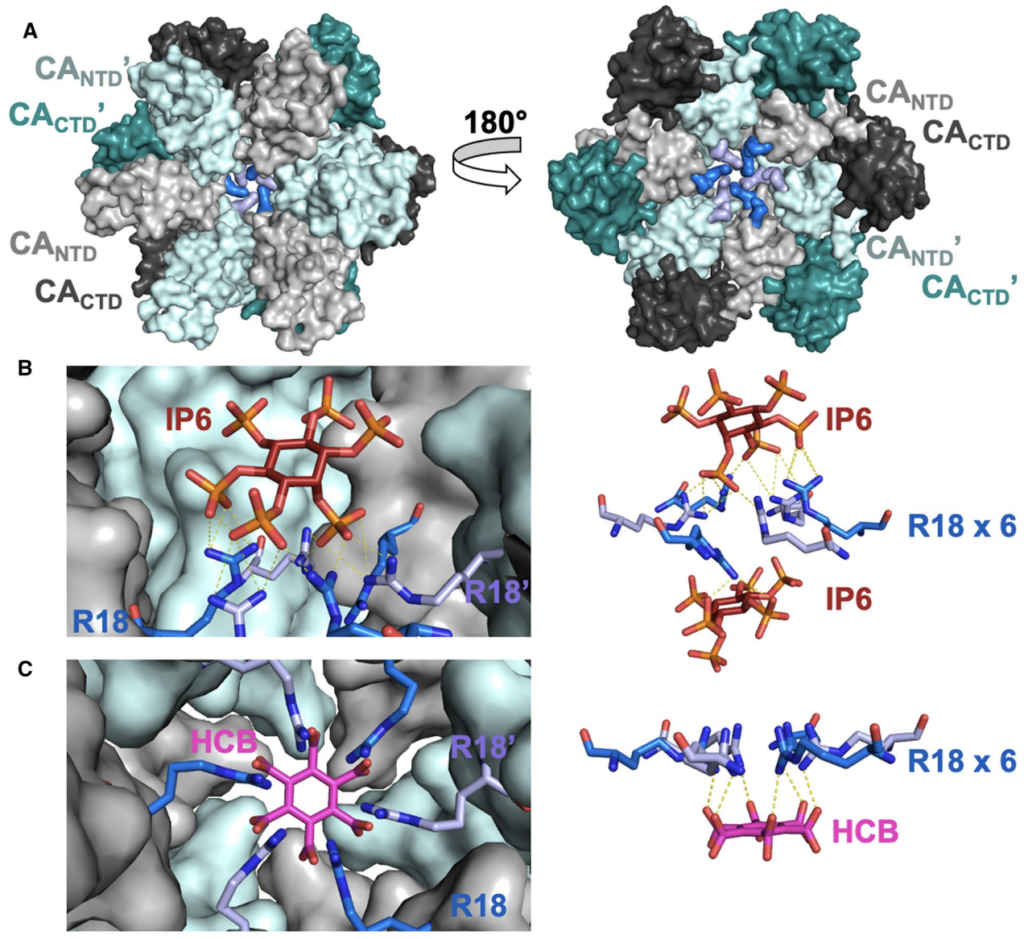
173. Targeting the HIV-1 and HBV Capsids, an EnCore. McFadden WM, Sarafianos SG. Viruses. 2023 Mar 31;15(4):896. doi: 10.3390/v15040896. PMID: 37112877.
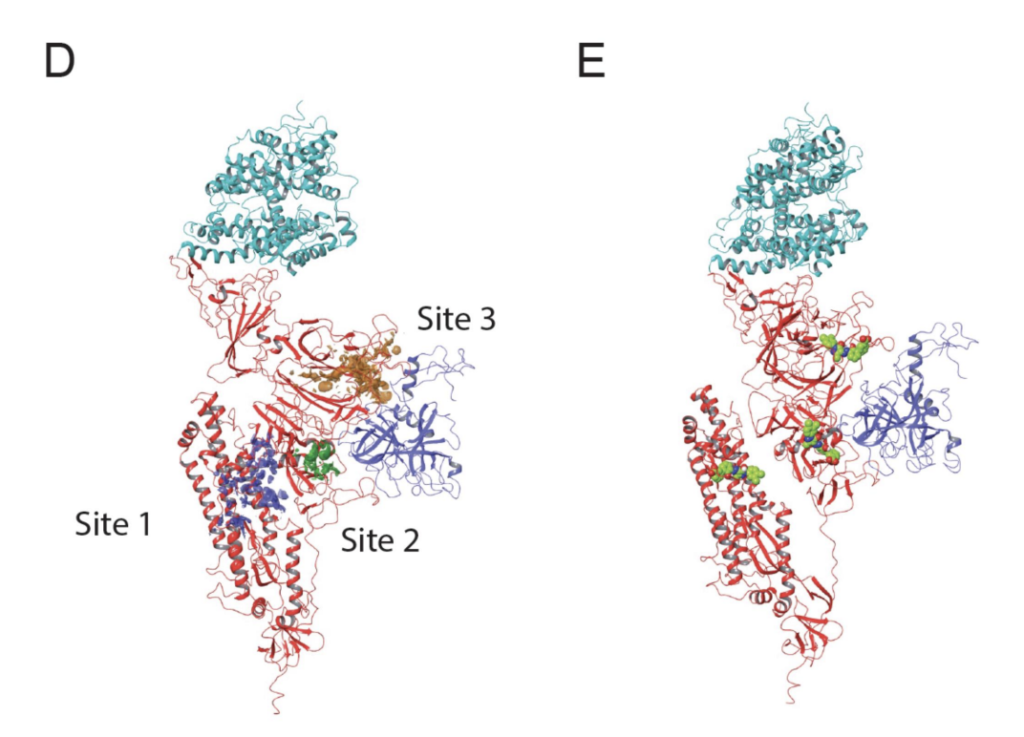
172. TMPRSS2 and SARS-CoV-2 SPIKE interaction assay for uHTS. Cicka D, Niu Q, Qui M, Qian K, Miller E, Fan D, Mo X, Ivanov AA, Sarafianos SG, Du Y, Fu H. J Mol Cell Biol. 2023 Mar 15:mjad017. doi: 10.1093/jmcb/mjad017. Epub ahead of print. PMID: 36921991.
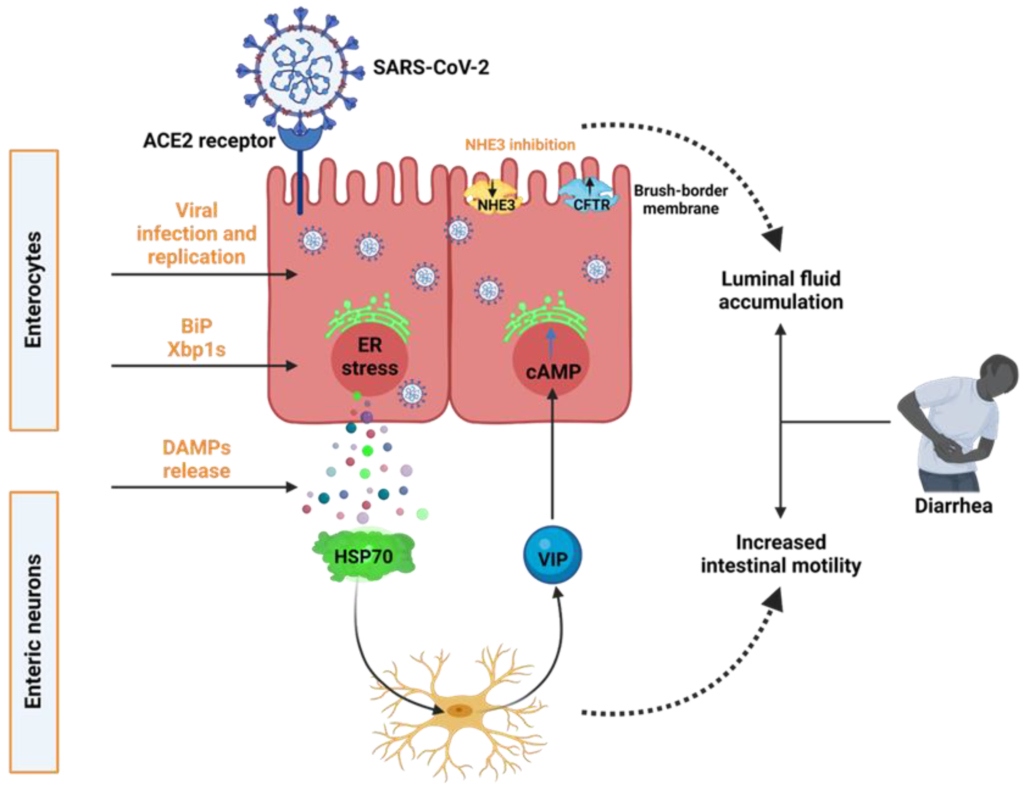
171. SARS-CoV-2 Induces Epithelial-Enteric Neuronal Crosstalk Stimulating VIP Release. Balasubramaniam A, Tedbury PR, Mwangi SM, Liu Y, Li G, Merlin D, Gracz AD, He P, Sarafianos SG, Srinivasan S. Biomolecules. 2023 Jan 20;13(2):207. doi: 10.3390/biom13020207. PMID: 36830577; PMCID: PMC9953368.
2022
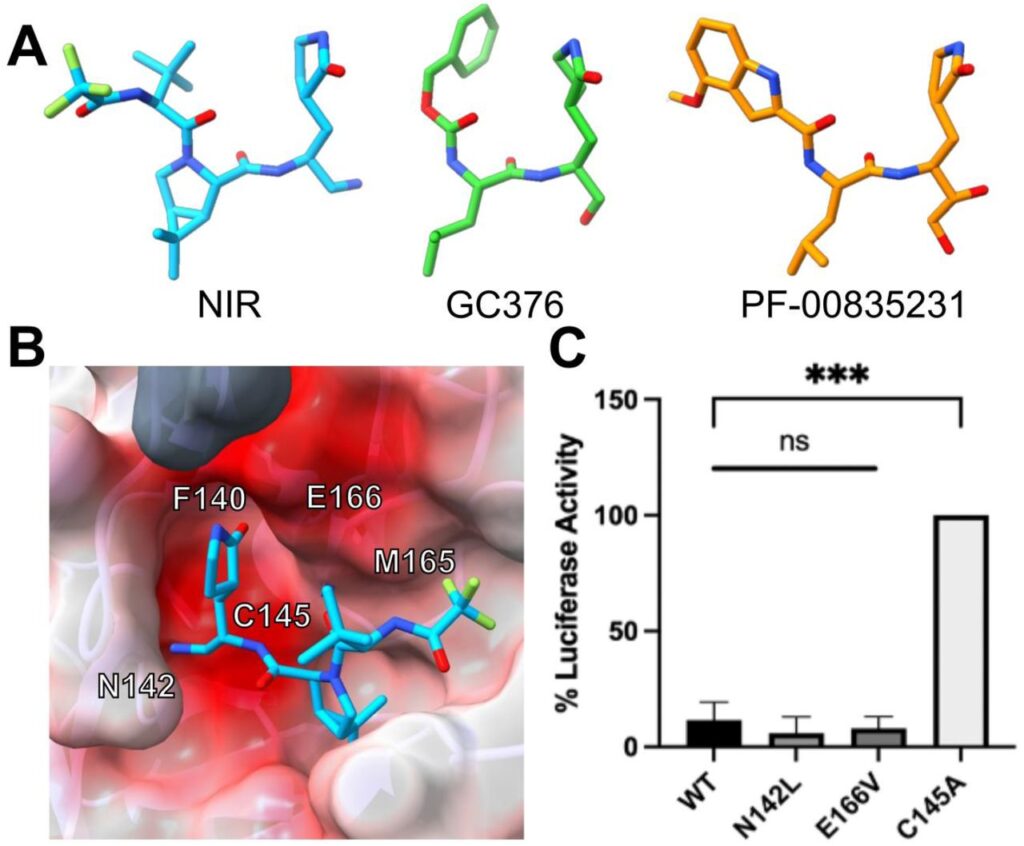
170. Nirmatrelvir Resistance in SARS-CoV-2 Omicron_BA.1 and WA1 Replicons and Escape Strategies. Lan S, Neilsen G, Slack RL, Cantara WA, Castaner AE, Lorson ZC, Lulkin N, Zhang H, Lee J, Cilento ME, Tedbury PR, Sarafianos SG. BioRxiv. 2022 December 31. doi: https://doi.org/10.1101/2022.12.31.522389.
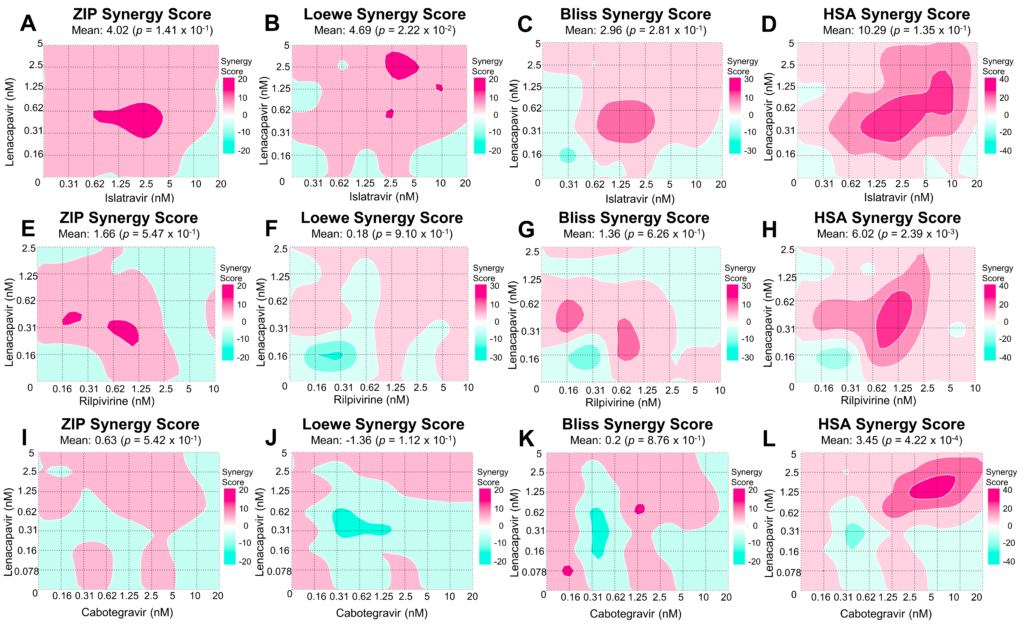
169. Drug Interactions in Lenacapavir-Based Long-Acting Antiviral Combinations. Cilento ME, Ong YT, Tedbury PR, Sarafianos SG. Viruses. 2022 May 31;14(6):1202. doi: https://doi.org/10.3390/v14061202. PMID:35746673; PMCID:PMC9229705
168. Marine Natural Products as Leads against SARS-CoV-2 Infection. Chhetri BK, Tedbury PR, Sweeney-Jones AM, Mani L, Soapi K, Manfredi C, Sorscher E, Sarafianos SG, Kubanek J. J Nat Prod. 2022 Mar 25;85(3):657-665. doi: 10.1021/acs.jnatprod.2c00015. Epub 2022 Mar 15. PMID: 35290044; PMCID: PMC8936055.
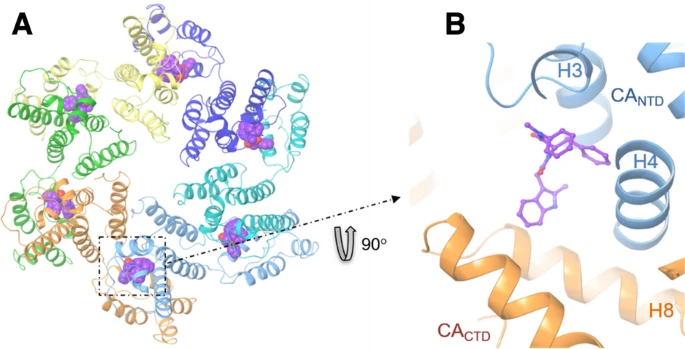
167. Structure-based virtual screening workflow to identify antivirals targeting HIV-1 capsid. Sun Q, Biswas A, Vijayan RSK, Craveur P, Forli S, Olson AJ, Castaner AE, Kirby KA, Sarafianos SG, Deng N, Levy R. J Comput Aided Mol Des. 2022 Mar 9:1–11. doi: 10.1007/s10822-022-00446-5. Epub ahead of print. PMID: 35262811; PMCID: PMC8904208.
166. TRIM5α Restriction of HIV-1-N74D Viruses in Lymphocytes Is Caused by a Loss of Cyclophilin A Protection. Selyutina A, Simons LM, Kirby KA, Bulnes-Ramos A, Hu P, Sarafianos SG, Hultquist JF, Diaz-Griffero F. Viruses. 2022 Feb 10;14(2):363. doi: 10.3390/v14020363. PMID: 35215956; PMCID: PMC8879423.
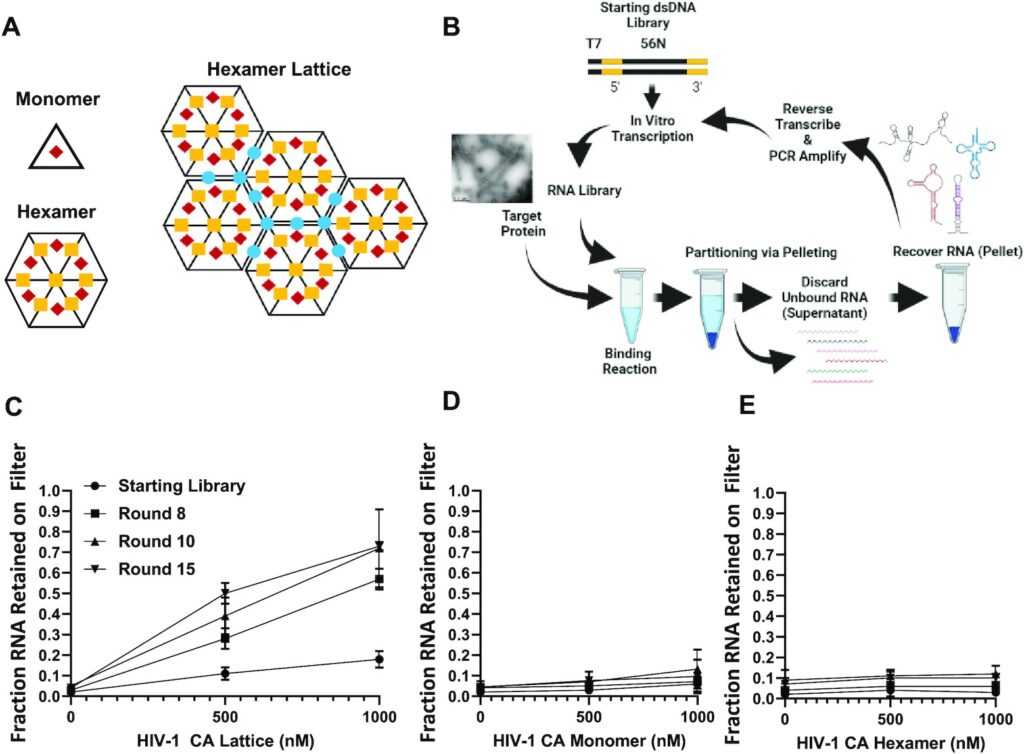
165. Selection and identification of an RNA aptamer that specifically binds the HIV-1 capsid lattice and inhibits viral replication. Gruenke PR, Aneja R, Welbourn S, Ukah OB, Sarafianos SG, Burke DH, Lange MJ. . Nucleic Acids Res. 2022 Feb 22;50(3):1701-1717. doi: 10.1093/nar/gkab1293. PMID: 35018437; PMCID: PMC8860611.
164. Specific mutations in the HIV-1 G-tract of the 3′-polypurine tract cause resistance to integrase strand transfer inhibitors. Hachiya A, Kubota M, Shigemi U, Ode H, Yokomaku Y, Kirby KA, Sarafianos SG, Iwatani Y. J Antimicrob Chemother. 2022 Feb 23;77(3):574-577. doi: 10.1093/jac/dkab448. PMID: 34894227; PMCID: PMC8865006.
2021
163. Rotten to the core: antivirals targeting the HIV-1 capsid core. McFadden WM, Snyder AA, Kirby KA, Tedbury PR, Raj M, Wang Z, Sarafianos SG. Retrovirology. 2021 Dec 22;18(1):41. doi: 10.1186/s12977-021-00583-z. PMID: 34937567; PMCID: PMC8693499.
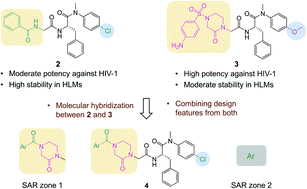
162. Potency and metabolic stability: a molecular hybrid case in the design of novel PF74-like small molecules targeting HIV-1 capsid protein. Sahani RL, Akther T, Cilento ME, Castaner AE, Zhang H, Kirby KA, Xie J, Sarafianos SG, Wang Z. RSC Med Chem. 2021 Sep 28;12(12):2031-2044. doi: 10.1039/d1md00292a. PMID: 35028563; PMCID: PMC8672819.
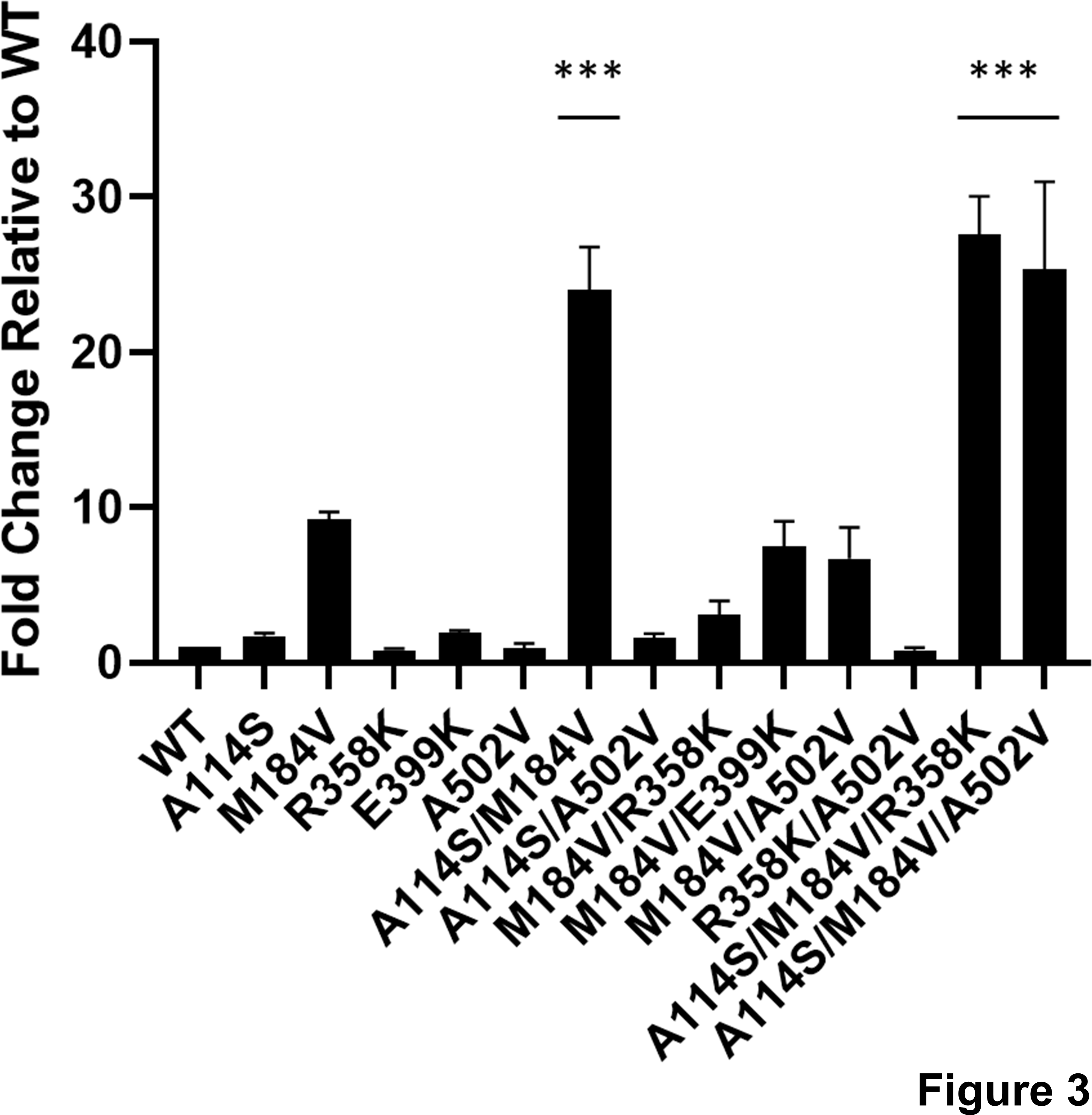
161. Development of Human Immunodeficiency Virus Type 1 Resistance to 4′-Ethynyl-2-Fluoro-2′-Deoxyadenosine (EFdA) Starting with Wild-Type or Nucleoside Reverse Transcriptase Inhibitor Resistant-Strains. Cilento ME, Reeve AB, Michailidis E, Ilina TV, Nagy E, Mitsuya H, Parniak MA, Tedbury PR, Sarafianos SG. Antimicrob Agents Chemother. 2021 Sep 13:AAC0116721. doi: 10.1128/AAC.01167-21. Online ahead of print. PMID: 34516245
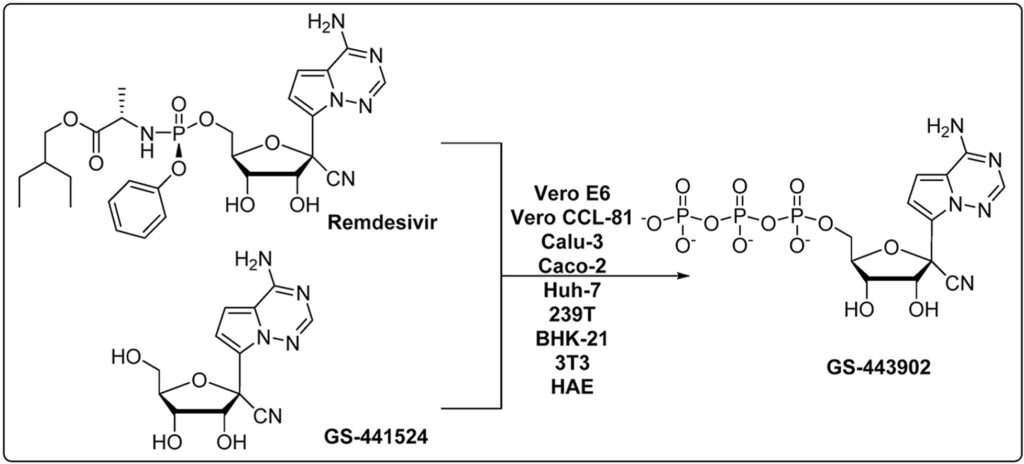
160. Comparison of anti-SARS-CoV-2 activity and intracellular metabolism of remdesivir and its parent nucleoside. Sijia Tao, Keivan Zandi, Leda Bassit, Yee Tsuey Ong, Kiran Verma, Peng Liu, Jessica A. Downs-Bowen, Tamara McBrayer, Julia C. Le Cher, James J. Kohler, Philip R. Tedbury, Baek Kim, Franck Amblard, Sarafianos SG, Raymond F. Schinazi. Curr Res Pharmacol Drug Discov. 2021;2:100045. doi: 10.1016/j.crphar.2021.100045. Epub 2021 Aug 12. PMID: 34870151; PMCID: PMC8357487.
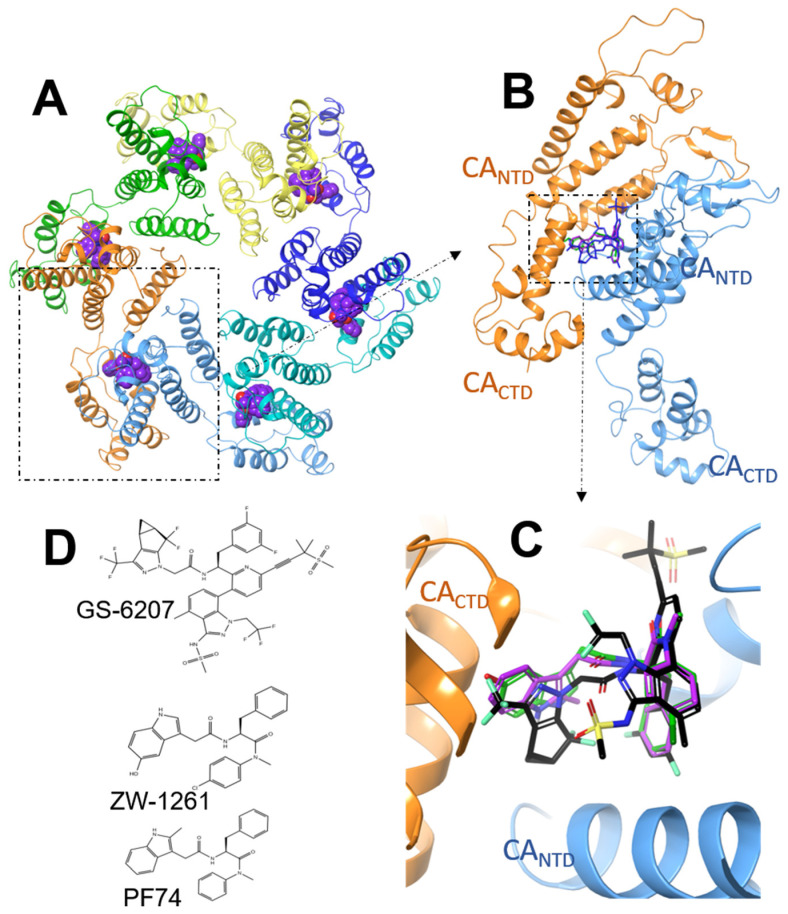
159. Molecular Dynamics Free Energy Simulations Reveal the Mechanism for the Antiviral Resistance of the M66I HIV-1 Capsid Mutation. Sun Q, Levy RM, Kirby KA, Wang Z, Sarafianos SG, Deng N. Viruses. 2021 May 15;13(5):920. doi: 10.3390/v13050920. PMID: 34063519 Free PMC article.
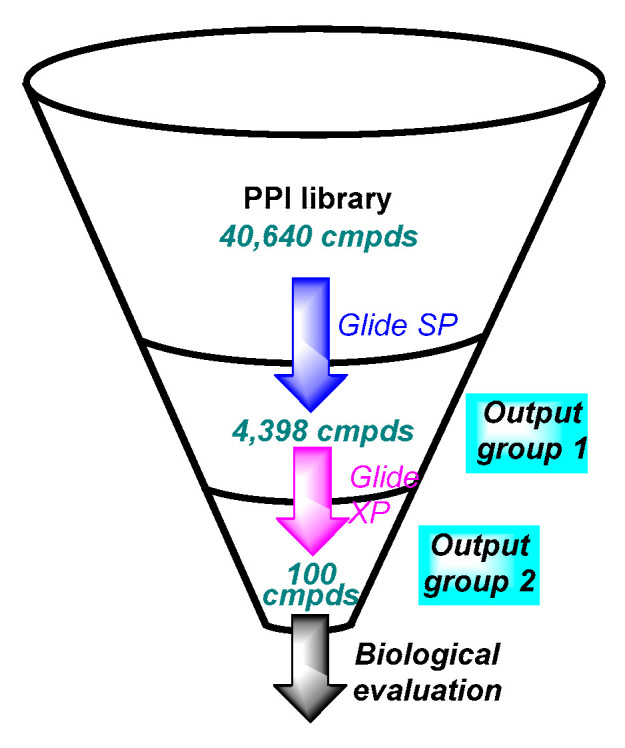
158. Discovery of New Small Molecule Hits as Hepatitis B Virus Capsid Assembly Modulators: Structure and Pharmacophore-Based Approaches. Senaweera S, Du H, Zhang H, Kirby KA, Tedbury PR, Xie J, Sarafianos SG, Wang Z. Viruses. 2021 Apr 27;13(5):770. doi: 10.3390/v13050770. PMID: 33925540 Free PMC article.
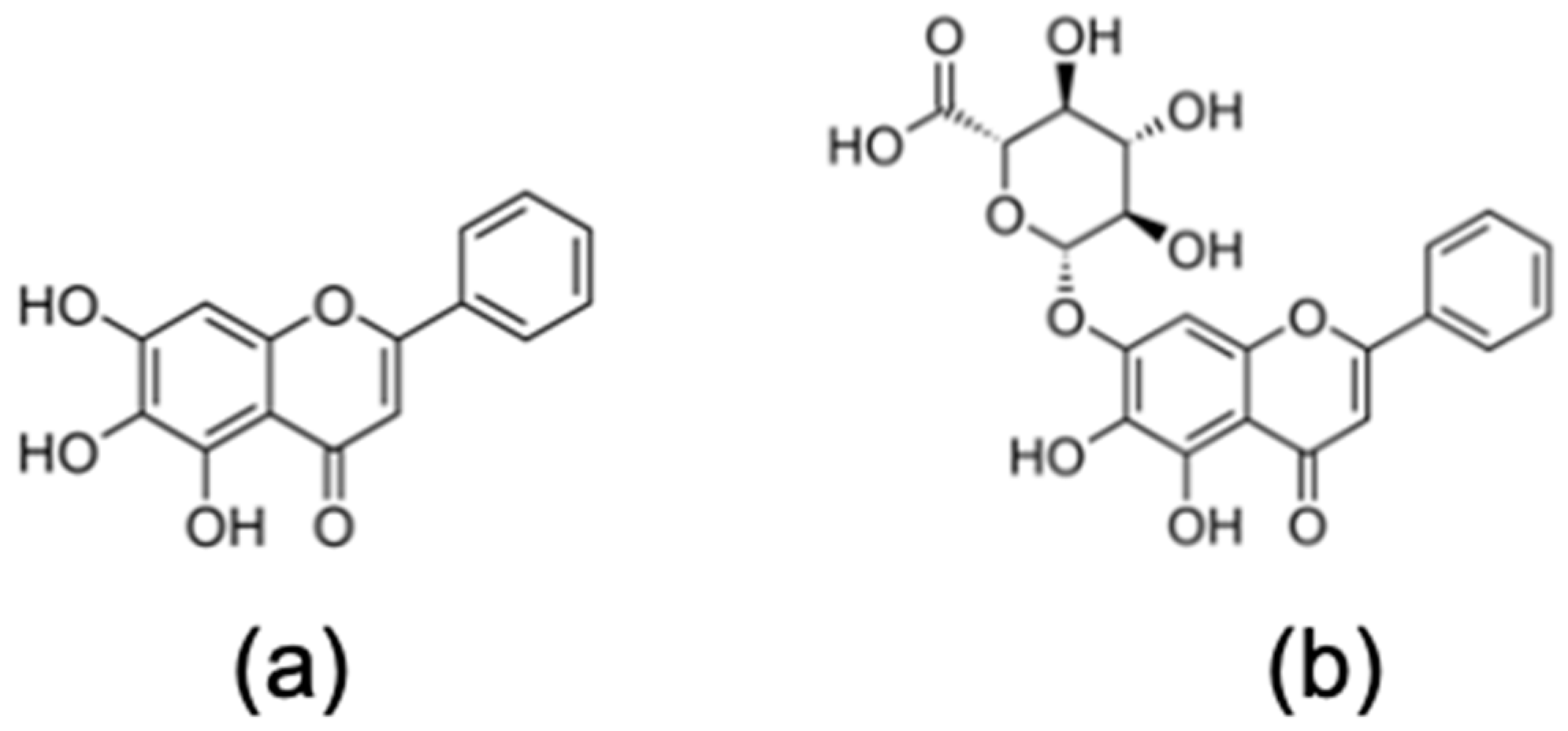
157. Baicalein and Baicalin Inhibit SARS-CoV-2 RNA-Dependent-RNA Polymerase. Zandi K, Musall K, Oo A, Cao D, Liang B, Hassandarvish P, Lan S, Slack RL, Kirby KA, Bassit L, Amblard F, Kim B, AbuBakar S, Sarafianos SG, Schinazi RF. Microorganisms. 2021 Apr 22;9(5):893. doi: 10.3390/microorganisms9050893. PMID: 33921971 Free PMC article.
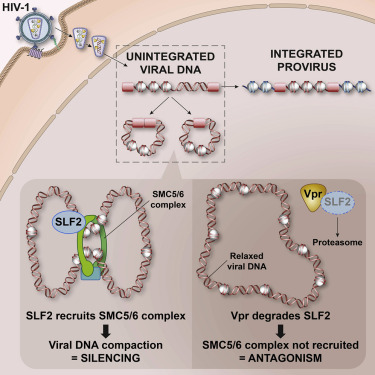
156. The SMC5/6 complex compacts and silences unintegrated HIV-1 DNA and is antagonized by Vpr. Dupont L, Bloor S, Williamson JC, Cuesta SM, Shah R, Teixeira-Silva A, Naamati A, Greenwood EJD, Sarafianos SG, Matheson NJ, Lehner PJ. Cell Host Microbe. 2021 Mar 26:S1931-3128(21)00101-3. doi: 10.1016/j.chom.2021.03.001. Online ahead of print. PMID: 33811831 Free article.
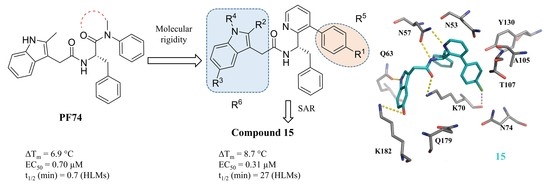
155. Design, Synthesis and Characterization of HIV-1 CA-Targeting Small Molecules: Conformational Restriction of PF74. Sahani RL, Diana-Rivero R, Vernekar SKV, Wang L, Du H, Zhang H, Castaner AE, Casey MC, Kirby KA, Tedbury PR, Xie J, Sarafianos SG, Wang Z. Viruses. 2021 Mar 15;13(3):479. doi: 10.3390/v13030479. PMID: 33804121 Free PMC article.
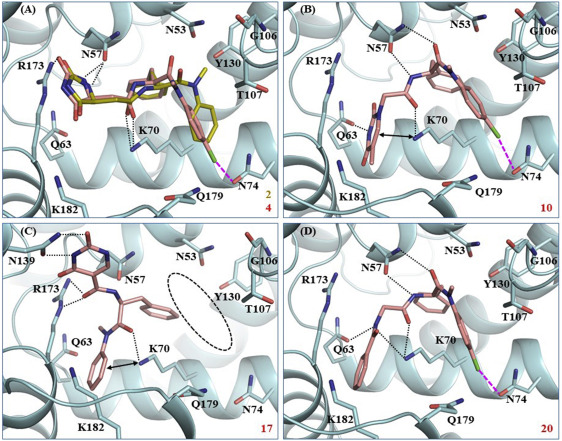
154. Novel PF74-like small molecules targeting the HIV-1 capsid protein: Balance of potency and metabolic stability. Wang L, Casey MC, Vernekar SKV, Sahani RL, Kirby KA, Du H, Zhang H, Tedbury PR, Xie J, Sarafianos SG, Wang Z. Acta Pharm Sin B. 2021 Mar;11(3):810-822. doi: 10.1016/j.apsb.2020.07.016. Epub 2020 Jul 31.PMID: 33777683 Free PMC article.
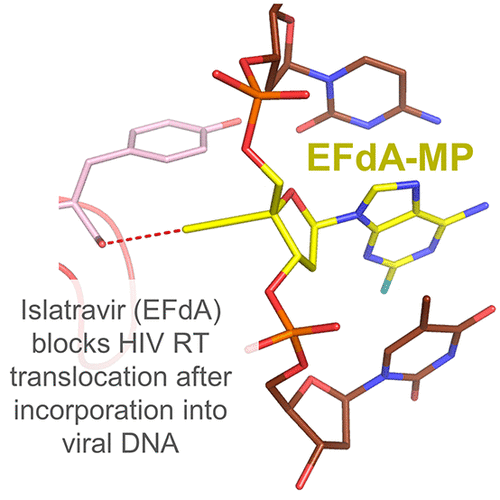
153. Avoiding Drug Resistance in HIV Reverse Transcriptase. Cilento ME, Kirby KA, Sarafianos SG. Chem Rev. 2021 Mar 24;121(6):3271-3296. doi: 10.1021/acs.chemrev.0c00967. Epub 2021 Jan 28. PMID: 33507067
2020
152. Single-cell Multiplexed Fluorescence Imaging to Visualize Viral Nucleic Acids and Proteins and Monitor HIV, HTLV, HBV, HCV, Zika Virus, and Influenza Infection. Shah R, Lan S, Puray-Chavez MN, Liu D, Tedbury PR, Sarafianos SG.J Vis Exp. 2020 Oct 29;(164):10.3791/61843. doi: 10.3791/61843.PMID: 33191939
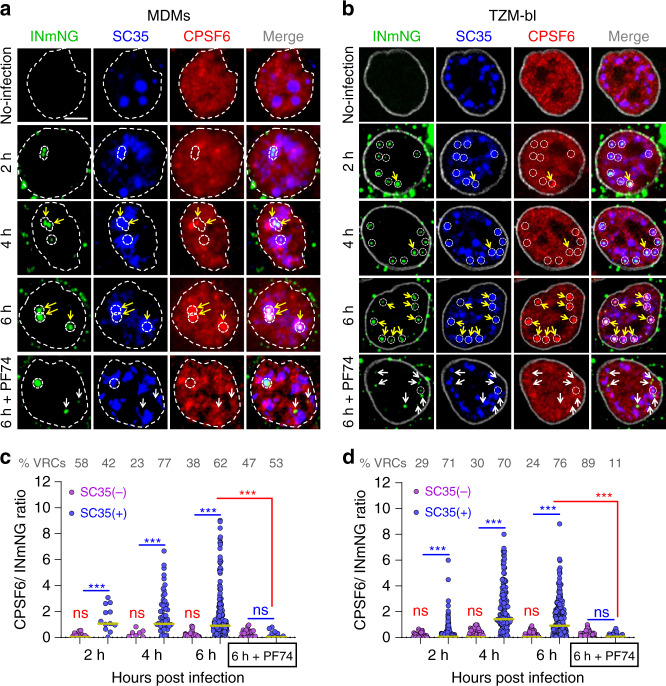
151. HIV-1 replication complexes accumulate in nuclear speckles and integrate into speckle-associated genomic domains. Francis AC, Marin M, Singh PK, Achuthan V, Prellberg MJ, Palermino-Rowland K, Lan S, Tedbury PR, Sarafianos SG, Engelman AN, Melikyan GB. Nat Commun. 2020 Jul 14;11(1):3505. doi: 10.1038/s41467-020-17256-8. PMID: 32665593
Publisher Correction: HIV-1 replication complexes accumulate in nuclear speckles and integrate into speckle-associated genomic domains. Francis AC, Marin M, Singh PK, Achuthan V, Prellberg MJ, Palermino-Rowland K, Lan S, Tedbury PR, Sarafianos SG, Engelman AN, Melikyan GB. Nat Commun. 2020 Nov 26;11(1):6165. doi: 10.1038/s41467-020-20152-w. PMID: 33244008 Free PMC article.
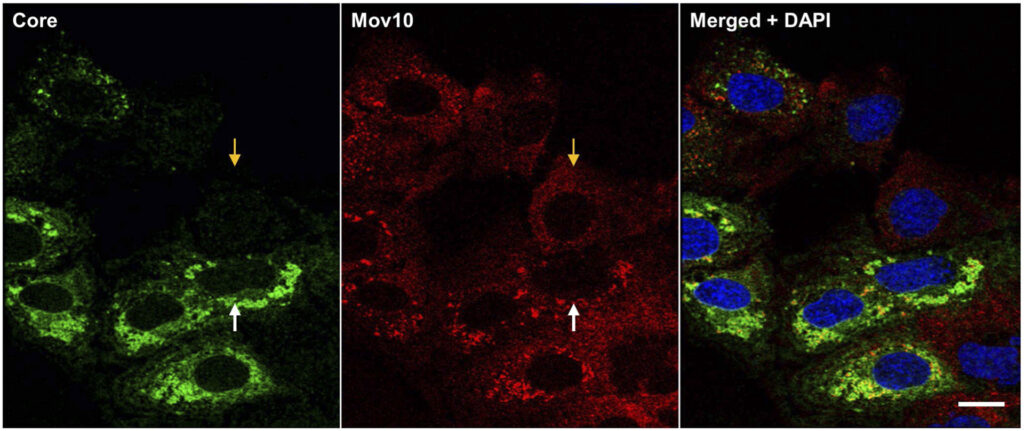
150. Effect of P-body component Mov10 on HCV virus production and infectivity. Liu D, Ndongwe TP, Puray-Chavez M, Casey MC, Izumi T, Pathak VK, Tedbury PR, Sarafianos SG. FASEB J. 2020 Jun 4. doi: 10.1096/fj.201800641R. Online ahead of print. PMID: 32496609
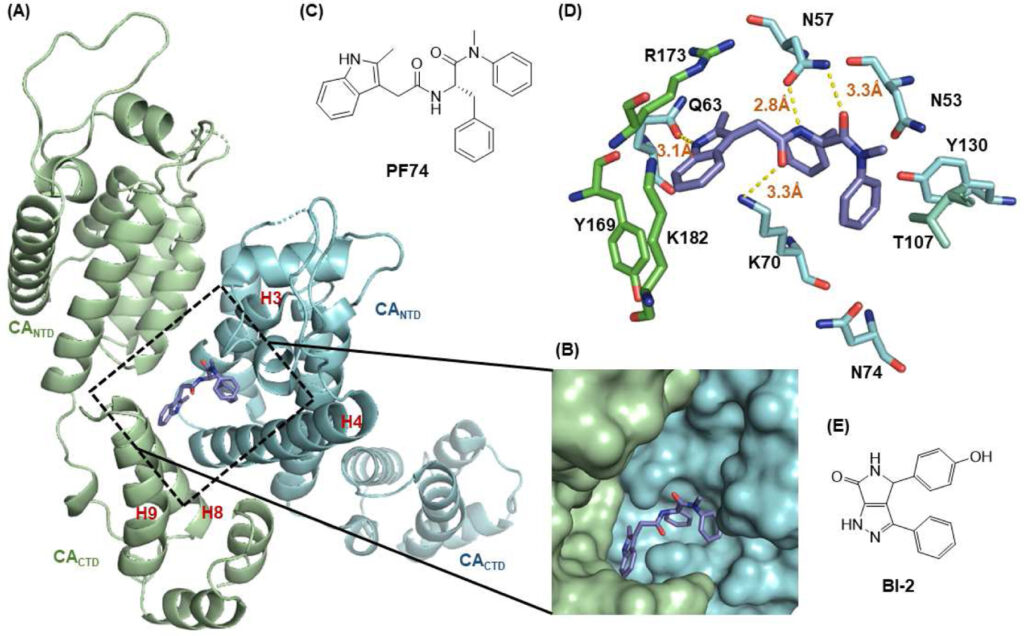
149. Chemical profiling of HIV-1 capsid-targeting antiviral PF74. Wang L, Casey MC, Vernekar SKV, Do HT, Sahani RL, Kirby KA, Du H, Hachiya A, Zhang H, Tedbury PR, Xie J, Sarafianos SG, Wang Z. Eur J Med Chem. 2020 Aug 15;200:112427. doi: 10.1016/j.ejmech.2020.112427. Epub 2020 May 12. PMID: 32438252
148. Feasibility of Known RNA Polymerase Inhibitors as Anti-SARS-CoV-2 Drugs. Neogi U, Hill KJ, Ambikan AT, Heng X, Quinn TP, Byrareddy SN, Sönnerborg A, Sarafianos SG, Singh K. Pathogens. 2020 Apr 26;9(5):320. doi: 10.3390/pathogens9050320. PMID: 32357471
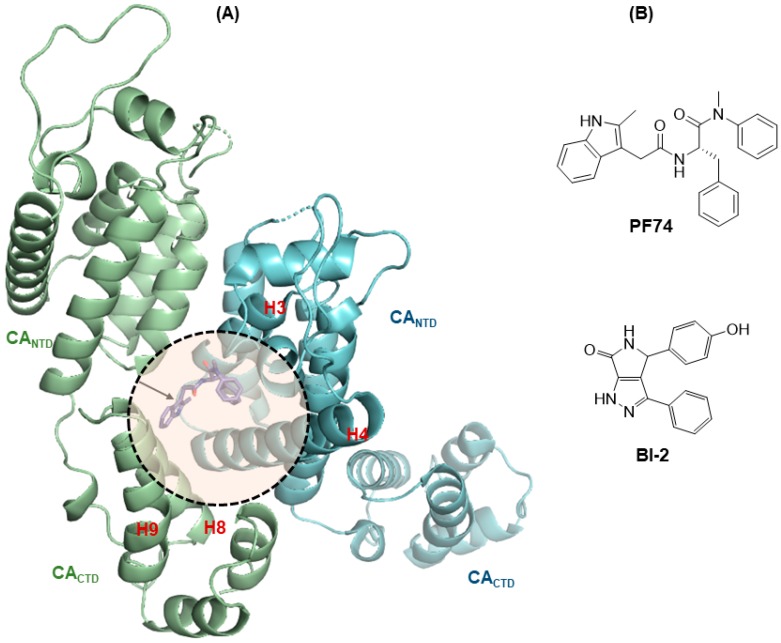
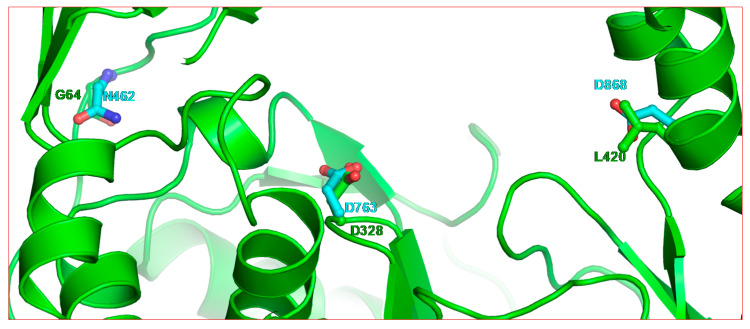
147. Toward Structurally Novel and Metabolically Stable HIV-1 Capsid-Targeting Small Molecules. Vernekar SKV, Sahani RL, Casey MC, Kankanala J, Wang L, Kirby KA, Du H, Zhang H, Tedbury PR, Xie J, Sarafianos SG, Wang Z. Viruses. 2020 Apr 16;12(4):452. doi: 10.3390/v12040452. PMID: 32316297
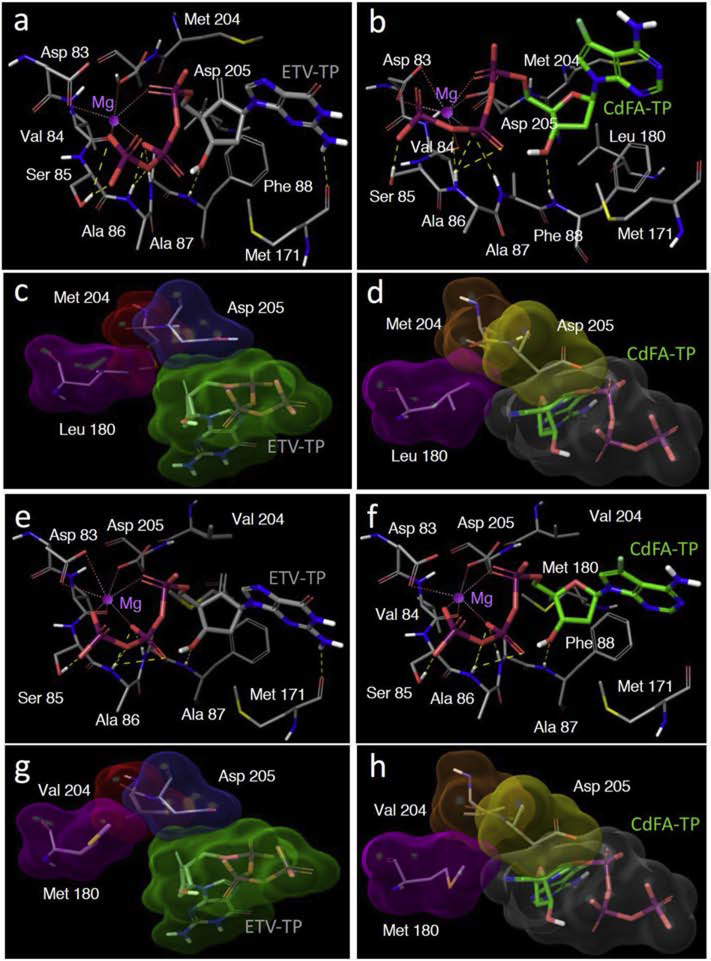
146. 7-Deaza-7-fluoro modification confers on 4′-cyano-nucleosides potent activity against entecavir/adefovir-resistant HBV variants and favorable safety. Hayashi S, Higashi-Kuwata N, Das D, Tomaya K, Yamada K, Murakami S, Venzon DJ, Hattori SI, Isogawa M, Sarafianos SG, Mitsuya H, Tanaka Y. Antiviral Res. 2020 Apr;176:104744. doi: 10.1016/j.antiviral.2020.104744. Epub 2020 Feb 18. PMID: 32084506
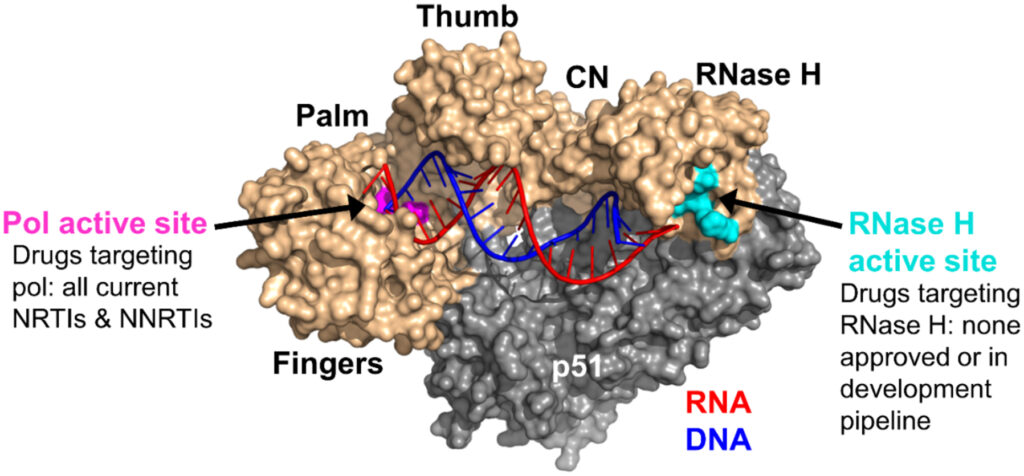
145. Cutting into the Substrate Dominance: Pharmacophore and Structure-Based Approaches toward Inhibiting Human Immunodeficiency Virus Reverse Transcriptase-Associated Ribonuclease H. Wang L, Sarafianos SG, Wang Z. Acc Chem Res. 2020 Jan 21;53(1):218-230. doi: 10.1021/acs.accounts.9b00450. Epub 2019 Dec 27. PMID: 31880912
- Visualization of Positive and Negative Sense Viral RNA for Probing the Mechanism of Direct-Acting Antivirals against Hepatitis C Virus. Liu D, Tedbury PR, Lan S, Huber AD, Puray-Chavez MN, Ji J, Michailidis E, Saeed M, Ndongwe TP, Bassit LC, Schinazi RF, Ralston R, Rice CM, Sarafianos SG. Viruses. 2019 Nov 8;11(11):1039. doi: 10.3390/v11111039. PMID: 31717338
- Determinants of Active-Site Inhibitor Interaction with HIV-1 RNase H. Xi Z, Wang Z, Sarafianos SG, Myshakina NS, Ishima R. ACS Infect Dis. 2019 Nov 8;5(11):1963-1974. doi: 10.1021/acsinfecdis.9b00300. Epub 2019 Oct 2. PMID: 31577424
- Glycosylated diphyllin as a broad-spectrum antiviral agent against Zika virus. Martinez-Lopez A, Persaud M, Chavez MP, Zhang H, Rong L, Liu S, Wang TT, Sarafianos SG, Diaz-Griffero F. EBioMedicine. 2019 Sep;47:269-283. doi: 10.1016/j.ebiom.2019.08.060. Epub 2019 Sep 6. PMID: 31501074
- Conformational Changes in HIV-1 Reverse Transcriptase that Facilitate Its Maturation. Slack RL, Ilina TV, Xi Z, Giacobbi NS, Kawai G, Parniak MA, Sarafianos SG, Sluis Cremer N, Ishima R. Structure. 2019 Oct 1;27(10):1581-1593.e3. doi: 10.1016/j.str.2019.08.004. Epub 2019 Aug 27. PMID: 31471129
- Effects of Moloney Leukemia Virus 10 Protein on Hepatitis B Virus Infection and Viral Replication. Puray-Chavez MN, Farghali MH, Yapo V, Huber AD, Liu D, Ndongwe TP, Casey MC, Laughlin TG, Hannink M, Tedbury PR, Sarafianos SG. Viruses. 2019 Jul 17;11(7):651. doi: 10.3390/v11070651. PMID: 31319455
- Long-Acting Anti-HIV Drugs Targeting HIV-1 Reverse Transcriptase and Integrase. Singh K, Sarafianos SG, Sönnerborg A. Pharmaceuticals (Basel). 2019 Apr 20;12(2):62. doi: 10.3390/ph12020062. PMID: 31010004
- Novel Intersubunit Interaction Critical for HIV-1 Core Assembly Defines a Potentially Targetable Inhibitor Binding Pocket. Craveur P, Gres AT, Kirby KA, Liu D, Hammond JA, Deng Y, Forli S, Goodsell DS, Williamson JR, Sarafianos SG, Olson AJ. mBio. 2019 Mar 12;10(2):e02858-18. doi: 10.1128/mBio.02858-18. PMID: 30862755
- Pharmacophore-based design of novel 3-hydroxypyrimidine-2,4-dione subtypes as inhibitors of HIV reverse transcriptase-associated RNase H: Tolerance of a nonflexible linker. Tang J, Do HT, Huber AD, Casey MC, Kirby KA, Wilson DJ, Kankanala J, Parniak MA, Sarafianos SG, Wang Z. Eur J Med Chem. 2019 Mar 15;166:390-399. doi: 10.1016/j.ejmech.2019.01.081. Epub 2019 Feb 2. PMID: 30739822
- CMCdG, a Novel Nucleoside Analog with Favorable Safety Features, Exerts Potent Activity against Wild-Type and Entecavir-Resistant Hepatitis B Virus. Higashi-Kuwata N, Hayashi S, Das D, Kohgo S, Murakami S, Hattori SI, Imoto S, Venzon DJ, Singh K, Sarafianos SG, Tanaka Y, Mitsuya H. Antimicrob Agents Chemother. 2019 Mar 27;63(4):e02143-18. doi: 10.1128/AAC.02143-18. Print 2019 Apr. PMID: 30670420
- 5-Aminothiophene-2,4-dicarboxamide analogues as hepatitis B virus capsid assembly effectors. Tang J, Huber AD, Pineda DL, Boschert KN, Wolf JJ, Kankanala J, Xie J, Sarafianos SG, Wang Z. Eur J Med Chem. 2019 Feb 15;164:179-192. doi: 10.1016/j.ejmech.2018.12.047. Epub 2018 Dec 21. PMID: 30594676
- Novel Hepatitis B Virus Capsid-Targeting Antiviral That Aggregates Core Particles and Inhibits Nuclear Entry of Viral Cores. Huber AD, Pineda DL, Liu D, Boschert KN, Gres AT, Wolf JJ, Coonrod EM, Tang J, Laughlin TG, Yang Q, Puray-Chavez MN, Ji J, Singh K, Kirby KA, Wang Z, Sarafianos SG. ACS Infect Dis. 2019 May 10;5(5):750-758. doi: 10.1021/acsinfecdis.8b00235. Epub 2019 Jan 14. PMID: 30582687
- Long-Acting Anti-HIV Drugs Targeting HIV-1 Reverse Transcriptase and Integrase. Singh K, Sarafianos SG, Sönnerborg A. Pharmaceuticals (Basel). 2019 Apr 20;12(2). pii: E62. doi: 10.3390/ph12020062. Review. PMID: 31010004
- CMCdG, a Novel Nucleoside Analog with Favorable Safety Features, Exerts Potent Activity against Wild-Type and Entecavir-Resistant Hepatitis B Virus. Higashi-Kuwata N, Hayashi S, Das D, Kohgo S, Murakami S, Hattori SI, Imoto S, Venzon DJ, Singh K, Sarafianos SG, Tanaka Y, Mitsuya H. Antimicrob Agents Chemother. 2019 Mar 27;63(4). pii: e02143-18. doi: 10.1128/AAC.02143-18. Print 2019 Apr. PMID: 30670420
- Pharmacophore-based design of novel 3-hydroxypyrimidine-2,4-dione subtypes as inhibitors of HIV reverse transcriptase-associated RNase H: Tolerance of a nonflexible linker. Tang J, Do HT, Huber AD, Casey MC, Kirby KA, Wilson DJ, Kankanala J, Parniak MA, Sarafianos SG, Wang Z. Eur J Med Chem. 2019 Mar 15;166:390-399. doi: 10.1016/j.ejmech.2019.01.081. Epub 2019 Feb 2. PMID: 30739822
- Novel Intersubunit Interaction Critical for HIV-1 Core Assembly Defines a Potentially Targetable Inhibitor Binding Pocket. Craveur P, Gres AT, Kirby KA, Liu D, Hammond JA, Deng Y, Forli S, Goodsell DS, Williamson JR, Sarafianos SG, Olson AJ. MBio. 2019 Mar 12;10(2). pii: e02858-18. doi: 10.1128/mBio.02858-18. PMID: 30862755
- Strain-specific effect on biphasic DNA binding by HIV-1 integrase. Hill KJ, Rogers LC, Njenda DT, Burke DH, Sarafianos SG, Sönnerborg A, Neogi U, Singh K. AIDS. 2019 Mar 1;33(3):588-592. doi: 10.1097/QAD.0000000000002078. PMID: 30475264
- 5-Aminothiophene-2,4-dicarboxamide analogues as hepatitis B virus capsid assembly effectors. Tang J, Huber AD, Pineda DL, Boschert KN, Wolf JJ, Kankanala J, Xie J, Sarafianos SG, Wang Z. Eur J Med Chem. 2019 Feb 15;164:179-192. doi: 10.1016/j.ejmech.2018.12.047. Epub 2018 Dec 21. PMID: 30594676
- Small Molecule Inhibitor that Stabilizes the Autoinhibited Conformation of the Oncogenic Tyrosine Phosphatase SHP2. Wu X, Xu G, Li X, Xu W, Li Q, Liu W, Kirby KA, Loh ML, Li J, Sarafianos SG, Qu CK. J Med Chem. 2019 Feb 14;62(3):1125-1137. doi: 10.1021/acs.jmedchem.8b00513. Epub 2018 Dec 5. PMID: 30457860
- The High Genetic Barrier of EFdA/MK-8591 Stems from Strong Interactions with the Active Site of Drug-Resistant HIV-1 Reverse Transcriptase. Takamatsu Y, Das D, Kohgo S, Hayashi H, Delino NS, Sarafianos SG, Mitsuya H, Maeda K. Cell Chem Biol. 2018 Oct 18;25(10):1268-1278.e3. doi: 10.1016/j.chembiol.2018.07.014. Epub 2018 Aug 30. PMID: 30174310
- Identification of a Structural Element in HIV-1 Gag Required for Virus Particle Assembly and Maturation. Novikova M, Adams LJ, Fontana J, Gres AT, Balasubramaniam M, Winkler DC, Kudchodkar SB, Soheilian F, Sarafianos SG, Steven AC, Freed EO. MBio. 2018 Oct 16;9(5). pii: e01567-18. doi: 10.1128/mBio.01567-18. PMID: 30327442
- Antiretroviral potency of 4′-ethnyl-2′-fluoro-2′-deoxyadenosine, tenofovir alafenamide and second-generation NNRTIs across diverse HIV-1 subtypes. Njenda DT, Aralaguppe SG, Singh K, Rao R, Sönnerborg A, Sarafianos SG, Neogi U. J Antimicrob Chemother. 2018 Oct 1;73(10):2721-2728. doi: 10.1093/jac/dky256. PMID: 30053052
- Visualization of HIV-1 RNA Transcription from Integrated HIV-1 DNA in Reactivated Latently Infected Cells. Ukah OB, Puray-Chavez M, Tedbury PR, Herschhorn A, Sodroski JG, Sarafianos SG. Viruses. 2018 Sep 30;10(10). pii: E534. doi: 10.3390/v10100534. PMID: 30274333
- Contribution of a Multifunctional Polymerase Region of Foot-and-Mouth Disease Virus to Lethal Mutagenesis. de la Higuera I, Ferrer-Orta C, Moreno E, de Ávila AI, Soria ME, Singh K, Caridi F, Sobrino F, Sarafianos SG, Perales C, Verdaguer N, Domingo E. J Virol. 2018 Sep 26;92(20). pii: e01119-18. doi: 10.1128/JVI.01119-18. Print 2018 Oct 15. PMID: 30068642
- Capsid-CPSF6 Interaction Licenses Nuclear HIV-1 Trafficking to Sites of Viral DNA Integration. Achuthan V, Perreira JM, Sowd GA, Puray-Chavez M, McDougall WM, Paulucci-Holthauzen A, Wu X, Fadel HJ, Poeschla EM, Multani AS, Hughes SH, Sarafianos SG, Brass AL, Engelman AN. Cell Host Microbe. 2018 Sep 12;24(3):392-404.e8. doi: 10.1016/j.chom.2018.08.002. Epub 2018 Aug 30. PMID: 30173955
- 6-Biphenylmethyl-3-hydroxypyrimidine-2,4-diones potently and selectively inhibited HIV reverse transcriptase-associated RNase H. Wang L, Tang J, Huber AD, Casey MC, Kirby KA, Wilson DJ, Kankanala J, Parniak MA, Sarafianos SG, Wang Z. Eur J Med Chem. 2018 Aug 5;156:680-691. doi: 10.1016/j.ejmech.2018.07.035. Epub 2018 Jul 17. PMID: 30031978
- 6-Arylthio-3-hydroxypyrimidine-2,4-diones potently inhibited HIV reverse transcriptase-associated RNase H with antiviral activity. Wang L, Tang J, Huber AD, Casey MC, Kirby KA, Wilson DJ, Kankanala J, Xie J, Parniak MA, Sarafianos SG, Wang Z. Eur J Med Chem. 2018 Aug 5;156:652-665. doi: 10.1016/j.ejmech.2018.07.039. Epub 2018 Jul 17. PMID: 30031976
- Structural Implications of Genotypic Variations in HIV-1 Integrase From Diverse Subtypes. Rogers L, Obasa AE, Jacobs GB, Sarafianos SG, Sönnerborg A, Neogi U, Singh K. Front Microbiol. 2018 Aug 2;9:1754. doi: 10.3389/fmicb.2018.01754. eCollection 2018. PMID: 30116231
- 4′-Ethynyl-2-fluoro-2′-deoxyadenosine, MK-8591: a novel HIV-1 reverse transcriptase translocation inhibitor. Markowitz M, Sarafianos SG. Curr Opin HIV AIDS. 2018 Jul;13(4):294-299. doi: 10.1097/COH.0000000000000467. Review. PMID: 29697468
- Effect of tRNA on the Maturation of HIV-1 Reverse Transcriptase. Ilina TV, Slack RL, Elder JH, Sarafianos SG, Parniak MA, Ishima R. J Mol Biol. 2018 Jun 22;430(13):1891-1900. doi: 10.1016/j.jmb.2018.02.027. Epub 2018 May 8. PMID: 29751015
- The Heteroaryldihydropyrimidine Bay 38-7690 Induces Hepatitis B Virus Core Protein Aggregates Associated with Promyelocytic Leukemia Nuclear Bodies in Infected Cells. Huber AD, Wolf JJ, Liu D, Gres AT, Tang J, Boschert KN, Puray-Chavez MN, Pineda DL, Laughlin TG, Coonrod EM, Yang Q, Ji J, Kirby KA, Wang Z, Sarafianos SG. mSphere 2018 Apr 18;3(2). pii: e00131-18. doi: 10.1128/mSphereDirect.00131-18. Print 2018 Apr 25. PMID: 29669885
- Ex-vivo antiretroviral potency of newer integrase strand transfer inhibitors cabotegravir and bictegravir in HIV type 1 non-B subtypes. Neogi U, Singh K, Aralaguppe SG, Rogers LC, Njenda DT, Sarafianos SG, Hejdeman B, Sönnerborg A. AIDS. 2018 Feb 20;32(4):469-476. doi: 10.1097/QAD.0000000000001726. PMID: 29239896
- Design, synthesis and biological evaluations of N-Hydroxy thienopyrimidine-2,4-diones as inhibitors of HIV reverse transcriptase-associated RNase H.Kankanala J, Kirby KA, Huber AD, Casey MC, Wilson DJ, Sarafianos SG, Wang Z. Eur J Med Chem. 2017 Dec 1;141:149-161. doi: 10.1016/j.ejmech.2017.09.054. Epub 2017 Sep 28. PMID:29031062
- Multiplex single-cell visualization of nucleic acids and protein during HIV infection.Puray-Chavez M, Tedbury PR, Huber AD, Ukah OB, Yapo V, Liu D, Ji J, Wolf JJ, Engelman AN, Sarafianos SG. Nat Commun. 2017 Dec 1;8(1):1882. doi: 10.1038/s41467-017-01693-z. PMID:29192235
- A 2-Hydroxyisoquinoline-1,3-Dione Active-Site RNase H Inhibitor Binds in Multiple Modes to HIV-1 Reverse Transcriptase.Kirby KA, Myshakina NA, Christen MT, Chen YL, Schmidt HA, Huber AD, Xi Z, Kim S, Rao RK, Kramer ST, Yang Q, Singh K, Parniak MA, Wang Z, Ishima R, Sarafianos SG. Antimicrob Agents Chemother. 2017 Sep 22;61(10). pii: e01351-17. doi: 10.1128/AAC.01351-17. Print 2017 Oct. PMID:28760905
- Impact of HIV-1 Integrase L74F and V75I Mutations in a Clinical Isolate on Resistance to Second-Generation Integrase Strand Transfer Inhibitors.Hachiya A, Kirby KA, Ido Y, Shigemi U, Matsuda M, Okazaki R, Imamura J, Sarafianos SG, Yokomaku Y, Iwatani Y. Antimicrob Agents Chemother. 2017 Jul 25;61(8). pii: e00315-17. doi: 10.1128/AAC.00315-17. Print 2017 Aug. PMID: 28533248
- Double-Winged 3-Hydroxypyrimidine-2,4-diones: Potent and Selective Inhibition against HIV-1 RNase H with Significant Antiviral Activity.Vernekar SKV, Tang J, Wu B, Huber AD, Casey MC, Myshakina N, Wilson DJ, Kankanala J, Kirby KA, Parniak MA, Sarafianos SG, Wang Z. J Med Chem. 2017 Jun 22;60(12):5045-5056. doi: 10.1021/acs.jmedchem.7b00440. Epub 2017 May 31. PMID: 28525279
- Synthesis, biological evaluation and molecular modeling of 2-Hydroxyisoquinoline-1,3-dione analogues as inhibitors of HIV reverse transcriptase associated ribonuclease H and polymerase. Tang J, Vernekar SKV, Chen YL, Miller L, Huber AD, Myshakina N, Sarafianos SG, Parniak MA, Wang Z. Eur J Med Chem. 2017 Jun 16;133:85-96. doi: 10.1016/j.ejmech.2017.03.059. Epub 2017 Mar 29. PMID: 28384548
- 3-Hydroxypyrimidine-2,4-Diones as Novel Hepatitis B Virus Antivirals Targeting the Viral Ribonuclease H. Huber AD, Michailidis E, Tang J, Puray-Chavez MN, Boftsi M, Wolf JJ, Boschert KN, Sheridan MA, Leslie MD, Kirby KA, Singh K, Mitsuya H, Parniak MA, Wang Z, Sarafianos SG. Antimicrob Agents Chemother. 2017 May 24;61(6). pii: e00245-17. doi: 10.1128/AAC.00245-17. Print 2017 Jun. PMID: 28320718
- Molecular and Functional Bases of Selection against a Mutation Bias in an RNA Virus. de la Higuera I, Ferrer-Orta C, de Ávila AI, Perales C, Sierra M, Singh K, Sarafianos SG, Dehouck Y, Bastolla U, Verdaguer N, Domingo E. Genome Biol Evol. 2017 May 1;9(5):1212-1228. doi: 10.1093/gbe/evx075. PMID: 28460010
- Exposing HIV’s weaknesses. Tedbury PR, Sarafianos SG. J Biol Chem. 2017 Apr 7;292(14):6027-6028. doi: 10.1074/jbc.H117.777714. Epub 2017 Apr 7. Review. PMID: 28389578
- 6-Cyclohexylmethyl-3-hydroxypyrimidine-2,4-dione as an inhibitor scaffold of HIV reverase transcriptase: Impacts of the 3-OH on inhibiting RNase H and polymerase. Tang J, Kirby KA, Huber AD, Casey MC, Ji J, Wilson DJ, Sarafianos SG, Wang Z. Eur J Med Chem. 2017 Mar 10;128:168-179. doi: 10.1016/j.ejmech.2017.01.041. Epub 2017 Jan 30. PMID: 28182989
- Small molecule inhibitors block Gas6-inducible TAM activation and tumorigenicity. Kimani SG, Kumar S, Bansal N, Singh K, Kholodovych V, Comollo T, Peng Y, Kotenko SV, Sarafianos SG, Bertino JR, Welsh WJ, Birge RB. Sci Rep. 2017 Mar 8;7:43908. doi: 10.1038/srep43908. PMID: 28272423
- Increased replication capacity following evolution of PYxE insertion in Gag-p6 is associated with enhanced virulence in HIV-1 subtype C from East Africa. Aralaguppe SG, Winner D, Singh K, Sarafianos SG, Quiñones-Mateu ME, Sönnerborg A, Neogi U. J Med Virol. 2017 Jan;89(1):106-111. doi: 10.1002/jmv.24610. Epub 2016 Jul 6. PMID: 27328744
- Structural basis of HIV inhibition by translocation-defective RT inhibitor 4′-ethynyl-2-fluoro-2′-deoxyadenosine (EFdA). Salie ZL, Kirby KA, Michailidis E, Marchand B, Singh K, Rohan LC, Kodama EN, Mitsuya H, Parniak MA, Sarafianos SG. Proc Natl Acad Sci U S A. 2016 Aug 16;113(33):9274-9. doi: 10.1073/pnas.1605223113. Epub 2016 Aug 3. PMID: 27489345
- 3-Hydroxypyrimidine-2,4-dione-5-N-benzylcarboxamides Potently Inhibit HIV-1 Integrase and RNase H. Wu B, Tang J, Wilson DJ, Huber AD, Casey MC, Ji J, Kankanala J, Xie J, Sarafianos SG, Wang Z. J Med Chem. 2016 Jul 14;59(13):6136-48. doi: 10.1021/acs.jmedchem.6b00040. Epub 2016 Jun 17. PMID: 27283261
- Design, Synthesis, and Biological Evaluations of Hydroxypyridonecarboxylic Acids as Inhibitors of HIV Reverse Transcriptase Associated RNase H. Kankanala J, Kirby KA, Liu F, Miller L, Nagy E, Wilson DJ, Parniak MA, Sarafianos SG, Wang Z. J Med Chem. 2016 May 26;59(10):5051-62. doi: 10.1021/acs.jmedchem.6b00465. Epub 2016 Apr 29. PMID: 27094954
- 3-Hydroxypyrimidine-2,4-diones as Selective Active Site Inhibitors of HIV Reverse Transcriptase-Associated RNase H: Design, Synthesis, and Biochemical Evaluations. Tang J, Liu F, Nagy E, Miller L, Kirby KA, Wilson DJ, Wu B, Sarafianos SG, Parniak MA, Wang Z. J Med Chem. 2016 Mar 24;59(6):2648-59. doi: 10.1021/acs.jmedchem.5b01879. Epub 2016 Mar 8. PMID: 26927866
- Factors influencing the efficacy of rilpivirine in HIV-1 subtype C in low- and middle-income countries. Neogi U, Häggblom A, Singh K, Rogers LC, Rao SD, Amogne W, Schülter E, Zazzi M, Arnold E, Sarafianos SG, Sönnerborg A. J Antimicrob Chemother. 2016 Feb;71(2):367-71. doi: 10.1093/jac/dkv359. Epub 2015 Oct 30. PMID: 26518047
- 4′-modified nucleoside analogs: potent inhibitors active against entecavir-resistant hepatitis B virus. Takamatsu Y, Tanaka Y, Kohgo S, Murakami S, Singh K, Das D, Venzon DJ, Amano M, Higashi-Kuwata N, Aoki M, Delino NS, Hayashi S, Takahashi S, Sukenaga Y, Haraguchi K, Sarafianos SG, Maeda K, Mitsuya H. Hepatology. 2015 Oct;62(4):1024-36. doi: 10.1002/hep.27962. Epub 2015 Aug 25. PMID: 26122273
- Vaginal Microbicide Film Combinations of Two Reverse Transcriptase Inhibitors, EFdA and CSIC, for the Prevention of HIV-1 Sexual Transmission. Zhang W, Hu M, Shi Y, Gong T, Dezzutti CS, Moncla B, Sarafianos SG, Parniak MA, Rohan LC. Pharm Res. 2015 Sep;32(9):2960-72. doi: 10.1007/s11095-015-1678-2. Epub 2015 Mar 21. PMID: 25794967
- STRUCTURAL VIROLOGY. X-ray crystal structures of native HIV-1 capsid protein reveal conformational variability. Gres AT, Kirby KA, KewalRamani VN, Tanner JJ, Pornillos O, Sarafianos SG. Science. 2015 Jul 3;349(6243):99-103. doi: 10.1126/science.aaa5936. Epub 2015 Jun 4. PMID: 26044298
- Oral administration of the nucleoside EFdA (4′-ethynyl-2-fluoro-2′-deoxyadenosine) provides rapid suppression of HIV viremia in humanized mice and favorable pharmacokinetic properties in mice and the rhesus macaque. Stoddart CA, Galkina SA, Joshi P, Kosikova G, Moreno ME, Rivera JM, Sloan B, Reeve AB, Sarafianos SG, Murphey-Corb M, Parniak MA. Antimicrob Agents Chemother. 2015 Jul;59(7):4190-8. doi: 10.1128/AAC.05036-14. Epub 2015 May 4. PMID: 25941222
- Multifunctionality of a picornavirus polymerase domain: nuclear localization signal and nucleotide recognition. Ferrer-Orta C, de la Higuera I, Caridi F, Sánchez-Aparicio MT, Moreno E, Perales C, Singh K, Sarafianos SG, Sobrino F, Domingo E, Verdaguer N. J Virol. 2015 Jul;89(13):6848-59. doi: 10.1128/JVI.03283-14. Epub 2015 Apr 22. PMID: 25903341
- Design, synthesis, biochemical, and antiviral evaluations of C6 benzyl and C6 biarylmethyl substituted 2-hydroxylisoquinoline-1,3-diones: dual inhibition against HIV reverse transcriptase-associated RNase H and polymerase with antiviral activities. Vernekar SK, Liu Z, Nagy E, Miller L, Kirby KA, Wilson DJ, Kankanala J, Sarafianos SG, Parniak MA, Wang Z. J Med Chem. 2015 Jan 22;58(2):651-64. doi: 10.1021/jm501132s. Epub 2014 Dec 30. PMID: 25522204
- CODAS syndrome is associated with mutations of LONP1, encoding mitochondrial AAA+ Lon protease. Strauss KA, Jinks RN, Puffenberger EG, Venkatesh S, Singh K, Cheng I, Mikita N, Thilagavathi J, Lee J, Sarafianos S, Benkert A, Koehler A, Zhu A, Trovillion V, McGlincy M, Morlet T, Deardorff M, Innes AM, Prasad C, Chudley AE, Lee IN, Suzuki CK. Am J Hum Genet. 2015 Jan 8;96(1):121-35. doi: 10.1016/j.ajhg.2014.12.003. PMID: 25574826
- Fast hepatitis C virus RNA elimination and NS5A redistribution by NS5A inhibitors studied by a multiplex assay approach. Liu D, Ji J, Ndongwe TP, Michailidis E, Rice CM, Ralston R, Sarafianos SG. Antimicrob Agents Chemother. 2015; 59(6):3482-92. doi: 10.1128/AAC.00223-15. Epub 2015 Apr 6. PMID: 25845863
- Structural basis of clade-specific HIV-1 neutralization by humanized anti-V3 monoclonal antibody KD-247. Kirby KA, Ong YT, Hachiya A, Laughlin TG, Chiang LA, Pan Y, Moran JL, Marchand B, Singh K, Gallazzi F, Quinn TP, Yoshimura K, Murakami T, Matsushita S, Sarafianos SG. FASEB J. 2015 Jan;29(1):70-80. doi: 10.1096/fj.14-252262. Epub 2014 Oct 28. PMID: 25351987
Remembering Professor Walter A. Scott. Sarafianos SG. Viruses. 2014 Oct 20;6(10):3873-4. doi: 10.3390/v6103873. PMID: 25333466
Antiviral drugs specific for coronaviruses in preclinical development. Adedeji AO, Sarafianos SG. Curr Opin Virol. 2014 Oct;8:45-53. doi: 10.1016/j.coviro.2014.06.002. Epub 2014 Jul 2. Review. PMID: 24997250
Drug resistance in non-B subtype HIV-1: impact of HIV-1 reverse transcriptase inhibitors. Singh K, Flores JA, Kirby KA, Neogi U, Sonnerborg A, Hachiya A, Das K, Arnold E, McArthur C, Parniak M, Sarafianos SG. Viruses. 2014 Sep 24;6(9):3535-62. doi: 10.3390/v6093535. Review. PMID: 25254383
4′-Ethynyl-2-fluoro-2′-deoxyadenosine (EFdA) inhibits HIV-1 reverse transcriptase with multiple mechanisms. Michailidis E, Huber AD, Ryan EM, Ong YT, Leslie MD, Matzek KB, Singh K, Marchand B, Hagedorn AN, Kirby KA, Rohan LC, Kodama EN, Mitsuya H, Parniak MA, Sarafianos SG. J Biol Chem. 2014 Aug 29;289(35):24533-48. doi: 10.1074/jbc.M114.562694. Epub 2014 Jun 26. PMID: 24970894
SAMHD1 has differential impact on the efficacies of HIV nucleoside reverse transcriptase inhibitors. Huber AD, Michailidis E, Schultz ML, Ong YT, Bloch N, Puray-Chavez MN, Leslie MD, Ji J, Lucas AD, Kirby KA, Landau NR, Sarafianos SG. Antimicrob Agents Chemother. 2014 Aug;58(8):4915-9. doi: 10.1128/AAC.02745-14. Epub 2014 May 27. PMID: 24867973
Evaluation of SSYA10-001 as a replication inhibitor of severe acute respiratory syndrome, mouse hepatitis, and Middle East respiratory syndrome coronaviruses. Adedeji AO, Singh K, Kassim A, Coleman CM, Elliott R, Weiss SR, Frieman MB, Sarafianos SG. Antimicrob Agents Chemother. 2014 Aug;58(8):4894-8. doi: 10.1128/AAC.02994-14. Epub 2014 May 19. PMID: 24841268
Preformulation studies of EFdA, a novel nucleoside reverse transcriptase inhibitor for HIV prevention. Zhang W, Parniak MA, Mitsuya H, Sarafianos SG, Graebing PW, Rohan LC. Drug Dev Ind Pharm. 2014 Aug;40(8):1101-11. doi: 10.3109/03639045.2013.809535. Epub 2013 Jul 10. PMID: 23841536
In vitro transport characteristics of EFdA, a novel nucleoside reverse transcriptase inhibitor using Caco-2 and MDCKII cell monolayers. Zhang W, Parniak MA, Sarafianos SG, Empey PE, Rohan LC. Eur J Pharmacol. 2014 Jun 5;732:86-95. doi: 10.1016/j.ejphar.2014.03.022. Epub 2014 Mar 29. PMID: 24690257
Probing the molecular mechanism of action of the HIV-1 reverse transcriptase inhibitor 4′-ethynyl-2-fluoro-2′-deoxyadenosine (EFdA) using pre-steady-state kinetics. Muftuoglu Y, Sohl CD, Mislak AC, Mitsuya H, Sarafianos SG, Anderson KS. Antiviral Res. 2014 Jun;106:1-4. doi: 10.1016/j.antiviral.2014.03.001. Epub 2014 Mar 12. PMID: 24632447
Development of a vaginal delivery film containing EFdA, a novel anti-HIV nucleoside reverse transcriptase inhibitor. Zhang W, Parniak MA, Sarafianos SG, Cost MR, Rohan LC. Int J Pharm. 2014 Jan 30;461(1-2):203-13. doi: 10.1016/j.ijpharm.2013.11.056. Epub 2013 Dec 9. PMID: 24333452
- Effects of substitutions at the 4′ and 2 positions on the bioactivity of 4′-ethynyl-2-fluoro-2′-deoxyadenosine. Kirby KA, Michailidis E, Fetterly TL, Steinbach MA, Singh K, Marchand B, Leslie MD, Hagedorn AN, Kodama EN, Marquez VE, Hughes SH, Mitsuya H, Parniak MA, Sarafianos SG. Antimicrob Agents Chemother. 2013 Dec;57(12):6254-64. doi: 10.1128/AAC.01703-13. Epub 2013 Oct 7. PMID: 24100493
- Future treatment strategies for novel Middle East respiratory syndrome coronavirus infection. Adedeji AO, Sarafianos SG. Future Med Chem. 2013 Dec;5(18):2119-22. doi: 10.4155/fmc.13.183. No abstract available. PMID: 24261888
- Structural requirements for RNA degradation by HIV-1 reverse transcriptase. Das K, Sarafianos SG, Arnold E. Nat Struct Mol Biol. 2013 Dec;20(12):1341-2. doi: 10.1038/nsmb.2725. No abstract available. PMID: 24304910
- Evaluation of Combinations of 4′-Ethynyl-2-Fluoro-2′-Deoxyadenosine with Clinically Used Antiretroviral Drugs. Hachiya A, Reeve AB, Marchand B, Michailidis E, Ong YT, Kirby KA, Leslie MD, Oka S, Kodama EN, Rohan LC, Mitsuya H, Parniak MA, Sarafianos SG. Antimicrob Agents Chemother. 2013 Sep;57(9):4554-4558. doi: 10.1128/AAC.00283-13. Epub 2013 Jun 24. PMID: 23796932
- HIV-1 resistance mechanism to an electrostatically constrained peptide fusion inhibitor that is active against T-20-resistant strains. Shimane K, Kawaji K, Miyamoto F, Oishi S, Watanabe K, Sakagami Y, Fujii N, Shimura K, Matsuoka M, Kaku M, Sarafianos SG, Kodama EN. Antimicrob Agents Chemother. 2013 Aug;57(8):4035-8. doi: 10.1128/AAC.00237-13. Epub 2013 May 20. PMID: 23689710
- Effect of mutations at position E138 in HIV-1 reverse transcriptase and their interactions with the M184I mutation on defining patterns of resistance to nonnucleoside reverse transcriptase inhibitors rilpivirine and etravirine. Xu HT, Colby-Germinario SP, Asahchop EL, Oliveira M, McCallum M, Schader SM, Han Y, Quan Y, Sarafianos SG, Wainberg MA. Antimicrob Agents Chemother. 2013 Jul;57(7):3100-9. doi: 10.1128/AAC.00348-13. Epub 2013 Apr 22. PMID: 23612196
- Novel inhibitors of severe acute respiratory syndrome coronavirus entry that act by three distinct mechanisms. Adedeji AO, Severson W, Jonsson C, Singh K, Weiss SR, Sarafianos SG. J Virol. 2013 Jul;87(14):8017-28. doi: 10.1128/JVI.00998-13. Epub 2013 May 15. PMID: 23678171
- Hypersusceptibility mechanism of Tenofovir-resistant HIV to EFdA. Michailidis E, Ryan EM, Hachiya A, Kirby KA, Marchand B, Leslie MD, Huber AD, Ong YT, Jackson JC, Singh K, Kodama EN, Mitsuya H, Parniak MA, Sarafianos SG. Retrovirology. 2013 Jun 24;10:65. doi: 10.1186/1742-4690-10-65. PMID: 23800377
- Repeated exposure to 5D9, an inhibitor of 3D polymerase, effectively limits the replication of foot-and-mouth disease virus in host cells. Rai DK, Schafer EA, Singh K, McIntosh MA, Sarafianos SG, Rieder E. Antiviral Res. 2013 Jun;98(3):380-5. doi: 10.1016/j.antiviral.2013.03.022. Epub 2013 Apr 8. PMID: 23578728
- Restriction fragment mass polymorphism (RFMP) analysis based on MALDI-TOF mass spectrometry for detecting antiretroviral resistance in HIV-1 infected patients. Lee JH, Hachiya A, Shin SK, Lee J, Gatanaga H, Oka S, Kirby KA, Ong YT, Sarafianos SG, Folk WR, Yoo W, Hong SP, Kim SO. Clin Microbiol Infect. 2013 Jun;19(6):E263-70. doi: 10.1111/1469-0691.12167. Epub 2013 Mar 11. PMID: 23480551
- Mechanism of resistance to S138A substituted enfuvirtide and its application to peptide design. Izumi K, Kawaji K, Miyamoto F, Shimane K, Shimura K, Sakagami Y, Hattori T, Watanabe K, Oishi S, Fujii N, Matsuoka M, Kaku M, Sarafianos SG, Kodama EN. Int J Biochem Cell Biol. 2013 Apr;45(4):908-15. doi: 10.1016/j.biocel.2013.01.015. Epub 2013 Jan 26. PMID: 23357451
- The hepatitis B virus ribonuclease H is sensitive to inhibitors of the human immunodeficiency virus ribonuclease H and integrase enzymes. Tavis JE, Cheng X, Hu Y, Totten M, Cao F, Michailidis E, Aurora R, Meyers MJ, Jacobsen EJ, Parniak MA, Sarafianos SG. PLoS Pathog. 2013 Jan;9(1):e1003125. doi: 10.1371/journal.ppat.1003125. Epub 2013 Jan 22. PMID: 23349632
- Effect of translocation defective reverse transcriptase inhibitors on the activity of N348I, a connection subdomain drug resistant HIV-1 reverse transcriptase mutant. Michailidis E, Singh K, Ryan EM, Hachiya A, Ong YT, Kirby KA, Marchand B, Kodama EN, Mitsuya H, Parniak MA, Sarafianos SG. Cell Mol Biol (Noisy-le-grand). 2012 Dec 22;58(1):187-95. PMID: 23273211
- Structural and biochemical basis for the difference in the helicase activity of two different constructs of SARS-CoV helicase. Adedeji AO, Singh K, Sarafianos SG. Cell Mol Biol (Noisy-le-grand). 2012 Dec 22;58(1):114-21. PMID: 23273200
- Preparation of biologically active single-chain variable antibody fragments that target the HIV-1 gp120 V3 loop. Ong YT, Kirby KA, Hachiya A, Chiang LA, Marchand B, Yoshimura K, Murakami T, Singh K, Matsushita S, Sarafianos SG. Cell Mol Biol (Noisy-le-grand). 2012 Dec 22;58(1):71-9. PMID: 23273194
- Biochemical mechanism of HIV-1 resistance to rilpivirine. Singh K, Marchand B, Rai DK, Sharma B, Michailidis E, Ryan EM, Matzek KB, Leslie MD, Hagedorn AN, Li Z, Norden PR, Hachiya A, Parniak MA, Xu HT, Wainberg MA, Sarafianos SG. J Biol Chem. 2012 Nov 2;287(45):38110-23. doi: 10.1074/jbc.M112.398180. Epub 2012 Sep 6. PMID: 22955279
- Inhibitors of HIV-1 Reverse Transcriptase-Associated Ribonuclease H Activity. Ilina T, Labarge K, Sarafianos SG, Ishima R, Parniak MA. Biology (Basel). 2012 Oct 19;1(3):521-41. doi: 10.3390/biology1030521. PMID: 23599900
- Wernicke’s encephalopathy complicating acute necrotic pancreatitis. Krokos N, Karakatsanis A, Sarafianos P, Koukou S. Am Surg. 2012 Sep;78(9):E390-2. No abstract available. PMID: 22964173
- Severe acute respiratory syndrome coronavirus replication inhibitor that interferes with the nucleic acid unwinding of the viral helicase. Adedeji AO, Singh K, Calcaterra NE, DeDiego ML, Enjuanes L, Weiss S, Sarafianos SG. Antimicrob Agents Chemother. 2012 Sep;56(9):4718-28. doi: 10.1128/AAC.00957-12. Epub 2012 Jun 25. PMID: 22733076
- Response of simian immunodeficiency virus to the novel nucleoside reverse transcriptase inhibitor 4′-ethynyl-2-fluoro-2′-deoxyadenosine in vitro and in vivo. Murphey-Corb M, Rajakumar P, Michael H, Nyaundi J, Didier PJ, Reeve AB, Mitsuya H, Sarafianos SG, Parniak MA. Antimicrob Agents Chemother. 2012 Sep;56(9):4707-12. doi: 10.1128/AAC.00723-12. Epub 2012 Jun 19. PMID: 22713337
- HIV-1 reverse transcriptase (RT) polymorphism 172K suppresses the effect of clinically relevant drug resistance mutations to both nucleoside and non-nucleoside RT inhibitors. Hachiya A, Marchand B, Kirby KA, Michailidis E, Tu X, Palczewski K, Ong YT, Li Z, Griffin DT, Schuckmann MM, Tanuma J, Oka S, Singh K, Kodama EN, Sarafianos SG. J Biol Chem. 2012 Aug 24;287(35):29988-99. doi: 10.1074/jbc.M112.351551. Epub 2012 Jul 2. PMID: 22761416
- Antiviral therapies: focus on hepatitis B reverse transcriptase. Michailidis E, Kirby KA, Hachiya A, Yoo W, Hong SP, Kim SO, Folk WR, Sarafianos SG. Int J Biochem Cell Biol. 2012 Jul;44(7):1060-71. doi: 10.1016/j.biocel.2012.04.006. Epub 2012 Apr 16. Review. PMID: 22531713
- Structural and inhibition studies of the RNase H function of xenotropic murine leukemia virus-related virus reverse transcriptase. Kirby KA, Marchand B, Ong YT, Ndongwe TP, Hachiya A, Michailidis E, Leslie MD, Sietsema DV, Fetterly TL, Dorst CA, Singh K, Wang Z, Parniak MA, Sarafianos SG. Antimicrob Agents Chemother. 2012 Apr;56(4):2048-61. doi: 10.1128/AAC.06000-11. Epub 2012 Jan 17. PMID: 22252812
- Mechanism of interaction of human mitochondrial DNA polymerase γ with the novel nucleoside reverse transcriptase inhibitor 4′-ethynyl-2-fluoro-2′-deoxyadenosine indicates a low potential for host toxicity. Sohl CD, Singh K, Kasiviswanathan R, Copeland WC, Mitsuya H, Sarafianos SG, Anderson KS. Antimicrob Agents Chemother. 2012 Mar;56(3):1630-4. doi: 10.1128/AAC.05729-11. Epub 2011 Dec 12. PMID: 22155823
- Biochemical, inhibition and inhibitor resistance studies of xenotropic murine leukemia virus-related virus reverse transcriptase. Ndongwe TP, Adedeji AO, Michailidis E, Ong YT, Hachiya A, Marchand B, Ryan EM, Rai DK, Kirby KA, Whatley AS, Burke DH, Johnson M, Ding S, Zheng YM, Liu SL, Kodama E, Delviks-Frankenberry KA, Pathak VK, Mitsuya H, Parniak MA, Singh K, Sarafianos SG. Nucleic Acids Res. 2012 Jan;40(1):345-59. doi: 10.1093/nar/gkr694. Epub 2011 Sep 8. PMID: 21908397
- Mechanism of nucleic acid unwinding by SARS-CoV helicase. Adedeji AO, Marchand B, Te Velthuis AJ, Snijder EJ, Weiss S, Eoff RL, Singh K, Sarafianos SG. PLoS One. 2012;7(5):e36521. doi: 10.1371/journal.pone.0036521. Epub 2012 May 15. PMID: 22615777
- Hepatitis B Virus genotypic differences map structurally close to NRTI resistance hot spots. Michailidis E, Singh K, Kirby KA, Hachiya A, Yoo W, Hong SP, Kim SO, Folk WR, Sarafianos SG. Int J Curr Chem. 2011 Oct;2(4):253-260. PMID: 22505793
- Broad-spectrum aptamer inhibitors of HIV reverse transcriptase closely mimic natural substrates. Ditzler MA, Bose D, Shkriabai N, Marchand B, Sarafianos SG, Kvaratskhelia M, Burke DH. Nucleic Acids Res. 2011 Oct;39(18):8237-47. doi: 10.1093/nar/gkr381. Epub 2011 Jul 3. PMID: 21727088
- Potent anti-HIV-1 activity of N-HR-derived peptides including a deep pocket-forming region without antagonistic effects on T-20. Izumi K, Watanabe K, Oishi S, Fujii N, Matsuoka M, Sarafianos SG, Kodama EN. Antivir Chem Chemother. 2011 Aug 23;22(1):51-5. doi: 10.3851/IMP1836. PMID: 21860071
- The sugar ring conformation of 4′-ethynyl-2-fluoro-2′-deoxyadenosine and its recognition by the polymerase active site of HIV reverse transcriptase. Kirby KA, Singh K, Michailidis E, Marchand B, Kodama EN, Ashida N, Mitsuya H, Parniak MA, Sarafianos SG. Cell Mol Biol (Noisy-le-grand). 2011 Feb 12;57(1):40-6. PMID: 21366961
- K70Q adds high-level tenofovir resistance to “Q151M complex” HIV reverse transcriptase through the enhanced discrimination mechanism. Hachiya A, Kodama EN, Schuckmann MM, Kirby KA, Michailidis E, Sakagami Y, Oka S, Singh K, Sarafianos SG. PLoS One. 2011 Jan 13;6(1):e16242. doi: 10.1371/journal.pone.0016242. PMID: 21249155
- Inhibitors of foot and mouth disease virus targeting a novel pocket of the RNA-dependent RNA polymerase. Durk RC, Singh K, Cornelison CA, Rai DK, Matzek KB, Leslie MD, Schafer E, Marchand B, Adedeji A, Michailidis E, Dorst CA, Moran J, Pautler C, Rodriguez LL, McIntosh MA, Rieder E, Sarafianos SG. PLoS One. 2010 Dec 21;5(12):e15049. doi: 10.1371/journal.pone.0015049. PMID: 21203539
- Resistance profiles of novel electrostatically constrained HIV-1 fusion inhibitors. Shimura K, Nameki D, Kajiwara K, Watanabe K, Sakagami Y, Oishi S, Fujii N, Matsuoka M, Sarafianos SG, Kodama EN. J Biol Chem. 2010 Dec 10;285(50):39471-80. doi: 10.1074/jbc.M110.145789. Epub 2010 Oct 11. PMID: 20937812
- The N348I mutation at the connection subdomain of HIV-1 reverse transcriptase decreases binding to nevirapine. Schuckmann MM, Marchand B, Hachiya A, Kodama EN, Kirby KA, Singh K, Sarafianos SG. J Biol Chem. 2010 Dec 3;285(49):38700-9. doi: 10.1074/jbc.M110.153783. Epub 2010 Sep 27. PMID: 20876531
- Divergent evolution in reverse transcriptase (RT) of HIV-1 group O and M lineages: impact on structure, fitness, and sensitivity to nonnucleoside RT inhibitors. Tebit DM, Lobritz M, Lalonde M, Immonen T, Singh K, Sarafianos S, Herchenröder O, Kräusslich HG, Arts EJ. J Virol. 2010 Oct;84(19):9817-30. doi: 10.1128/JVI.00991-10. Epub 2010 Jul 14. PMID: 20631150
- Structural basis of HIV-1 resistance to AZT by excision. Tu X, Das K, Han Q, Bauman JD, Clark AD Jr, Hou X, Frenkel YV, Gaffney BL, Jones RA, Boyer PL, Hughes SH, Sarafianos SG, Arnold E. Nat Struct Mol Biol. 2010 Oct;17(10):1202-9. doi: 10.1038/nsmb.1908. Epub 2010 Sep 19. PMID: 20852643
- Rev-derived peptides inhibit HIV-1 replication by antagonism of Rev and a co-receptor, CXCR4. Shimane K, Kodama EN, Nakase I, Futaki S, Sakurai Y, Sakagami Y, Li X, Hattori T, Sarafianos SG, Matsuoka M. Int J Biochem Cell Biol. 2010 Sep;42(9):1482-8. doi: 10.1016/j.biocel.2010.05.005. Epub 2010 May 16. PMID: 20580677
- Reverse transcriptase in motion: conformational dynamics of enzyme-substrate interactions. Götte M, Rausch JW, Marchand B, Sarafianos S, Le Grice SF. Biochim Biophys Acta. 2010 May;1804(5):1202-12. doi: 10.1016/j.bbapap.2009.07.020. Epub 2009 Aug 7. Review. PMID: 19665597
- Structural Aspects of Drug Resistance and Inhibition of HIV-1 Reverse Transcriptase. Singh K, Marchand B, Kirby KA, Michailidis E, Sarafianos SG. Viruses. 2010 Feb 11;2(2):606-638. PMID: 20376302
- The mutation T477A in HIV-1 reverse transcriptase (RT) restores normal proteolytic processing of RT in virus with Gag-Pol mutated in the p51-RNH cleavage site. Abram ME, Sarafianos SG, Parniak MA. Retrovirology. 2010 Feb 1;7:6. doi: 10.1186/1742-4690-7-6. PMID: 20122159
- Mechanism of inhibition of HIV-1 reverse transcriptase by 4′-Ethynyl-2-fluoro-2′-deoxyadenosine triphosphate, a translocation-defective reverse transcriptase inhibitor. Michailidis E, Marchand B, Kodama EN, Singh K, Matsuoka M, Kirby KA, Ryan EM, Sawani AM, Nagy E, Ashida N, Mitsuya H, Parniak MA, Sarafianos SG. J Biol Chem. 2009 Dec 18;284(51):35681-91. doi: 10.1074/jbc.M109.036616. PMID: 19837673
- Structural basis for the role of the K65R mutation in HIV-1 reverse transcriptase polymerization, excision antagonism, and tenofovir resistance. Das K, Bandwar RP, White KL, Feng JY, Sarafianos SG, Tuske S, Tu X, Clark AD Jr, Boyer PL, Hou X, Gaffney BL, Jones RA, Miller MD, Hughes SH, Arnold E. J Biol Chem. 2009 Dec 11;284(50):35092-100. doi: 10.1074/jbc.M109.022525. Epub 2009 Oct 7. PMID: 19812032
- Synthesis of Boranoate, Selenoate, and Thioate Analogs of AZTp(4)A and Ap(4)A. Han Q, Sarafianos SG, Arnold E, Parniak MA, Gaffney BL, Jones RA. Tetrahedron. 2009 Sep 19;65(38):7915-7920. PMID: 20625456
- Clinical relevance of substitutions in the connection subdomain and RNase H domain of HIV-1 reverse transcriptase from a cohort of antiretroviral treatment-naïve patients. Hachiya A, Shimane K, Sarafianos SG, Kodama EN, Sakagami Y, Negishi F, Koizumi H, Gatanaga H, Matsuoka M, Takiguchi M, Oka S. Antiviral Res. 2009 Jun;82(3):115-21. doi: 10.1016/j.antiviral.2009.02.189. Epub 2009 Feb 21. PMID: 19428602
- SC29EK, a peptide fusion inhibitor with enhanced alpha-helicity, inhibits replication of human immunodeficiency virus type 1 mutants resistant to enfuvirtide. Naito T, Izumi K, Kodama E, Sakagami Y, Kajiwara K, Nishikawa H, Watanabe K, Sarafianos SG, Oishi S, Fujii N, Matsuoka M. Antimicrob Agents Chemother. 2009 Mar;53(3):1013-8. doi: 10.1128/AAC.01211-08. Epub 2008 Dec 29. PMID: 19114674
- Design of peptide-based inhibitors for human immunodeficiency virus type 1 strains resistant to T-20. Izumi K, Kodama E, Shimura K, Sakagami Y, Watanabe K, Ito S, Watabe T, Terakawa Y, Nishikawa H, Sarafianos SG, Kitaura K, Oishi S, Fujii N, Matsuoka M. J Biol Chem. 2009 Feb 20;284(8):4914-20. doi: 10.1074/jbc.M807169200. Epub 2008 Dec 10. PMID: 19073606
- Structure and function of HIV-1 reverse transcriptase: molecular mechanisms of polymerization and inhibition. Sarafianos SG, Marchand B, Das K, Himmel DM, Parniak MA, Hughes SH, Arnold E. J Mol Biol. 2009 Jan 23;385(3):693-713. doi: 10.1016/j.jmb.2008.10.071. Epub 2008 Nov 3. Review. PMID: 19022262
- Biochemistry. RT slides home… Sarafianos SG, Arnold E. Science. 2008 Nov 14;322(5904):1059-60. doi: 10.1126/science.1167454. No abstract available. PMID: 19008434
- The RNA polymerase “switch region” is a target for inhibitors. Mukhopadhyay J, Das K, Ismail S, Koppstein D, Jang M, Hudson B, Sarafianos S, Tuske S, Patel J, Jansen R, Irschik H, Arnold E, Ebright RH. Cell. 2008 Oct 17;135(2):295-307. doi: 10.1016/j.cell.2008.09.033. PMID: 18957204
- Molecular biology: an HIV secret uncovered. Arnold E, Sarafianos SG. Nature. 2008 May 8;453(7192):169-70. doi: 10.1038/453169b. No abstract available. PMID: 18464731
- Amino acid mutation N348I in the connection subdomain of human immunodeficiency virus type 1 reverse transcriptase confers multiclass resistance to nucleoside and nonnucleoside reverse transcriptase inhibitors. Hachiya A, Kodama EN, Sarafianos SG, Schuckmann MM, Sakagami Y, Matsuoka M, Takiguchi M, Gatanaga H, Oka S. J Virol. 2008 Apr;82(7):3261-70. doi: 10.1128/JVI.01154-07. Epub 2008 Jan 23. PMID: 18216099
- Identification and characterization of coumestans as novel HCV NS5B polymerase inhibitors. Kaushik-Basu N, Bopda-Waffo A, Talele TT, Basu A, Costa PR, da Silva AJ, Sarafianos SG, Noël F. Nucleic Acids Res. 2008 Mar;36(5):1482-96. doi: 10.1093/nar/gkm1178. Epub 2008 Jan 18. PMID: 18203743
- 2′-deoxy-4′-C-ethynyl-2-halo-adenosines active against drug-resistant human immunodeficiency virus type 1 variants. Kawamoto A, Kodama E, Sarafianos SG, Sakagami Y, Kohgo S, Kitano K, Ashida N, Iwai Y, Hayakawa H, Nakata H, Mitsuya H, Arnold E, Matsuoka M. Int J Biochem Cell Biol. 2008;40(11):2410-20. doi: 10.1016/j.biocel.2008.04.007. Epub 2008 Apr 11. PMID: 18487070
- Synthesis of conformationally locked carbocyclic nucleoside phosphonates to probe the active site of HIV-1 RT. Saneyoshi H, Vu BC, Hughes SH, Boyer PL, Sarafianos SG, Marquez VE. Nucleic Acids Symp Ser (Oxf). 2008;(52):623-4. doi: 10.1093/nass/nrn315. PMID: 18776534
- Synthesis of AZTpSpCX2ppSA and AZTpSpCX2ppSAZT: hydrolysis-resistant potential inhibitors of the AZT excision reaction of HIV-1 RT. Han Q, Sarafianos SG, Arnold E, Parniak MA, Gaffney BL, Jones RA. Org Lett. 2007 Dec 6;9(25):5243-6. Epub 2007 Nov 8. PMID: 17988141
- 3′-Azido-3′-deoxythymidine-(5′)-tetraphospho-(5′)-adenosine, the product of ATP-mediated excision of chain-terminating AZTMP, is a potent chain-terminating substrate for HIV-1 reverse transcriptase. Dharmasena S, Pongracz Z, Arnold E, Sarafianos SG, Parniak MA. Biochemistry. 2007 Jan 23;46(3):828-36. PMID: 17223704
- Crystal structures of clinically relevant Lys103Asn/Tyr181Cys double mutant HIV-1 reverse transcriptase in complexes with ATP and non-nucleoside inhibitor HBY 097. Das K, Sarafianos SG, Clark AD Jr, Boyer PL, Hughes SH, Arnold E. J Mol Biol. 2007 Jan 5;365(1):77-89. Epub 2006 Sep 15. PMID: 17056061
- HIV-1 reverse transcriptase structure with RNase H inhibitor dihydroxy benzoyl naphthyl hydrazone bound at a novel site. Himmel DM, Sarafianos SG, Dharmasena S, Hossain MM, McCoy-Simandle K, Ilina T, Clark AD Jr, Knight JL, Julias JG, Clark PK, Krogh-Jespersen K, Levy RM, Hughes SH, Parniak MA, Arnold E. ACS Chem Biol. 2006 Dec 20;1(11):702-12. PMID: 17184135
- Combining mutations in HIV-1 reverse transcriptase with mutations in the HIV-1 polypurine tract affects RNase H cleavages involved in PPT utilization. McWilliams MJ, Julias JG, Sarafianos SG, Alvord WG, Arnold E, Hughes SH. Virology. 2006 May 10;348(2):378-88. Epub 2006 Feb 10. PMID: 16473384
- Structural determinants of slippage-mediated mutations by human immunodeficiency virus type 1 reverse transcriptase. Hamburgh ME, Curr KA, Monaghan M, Rao VR, Tripathi S, Preston BD, Sarafianos S, Arnold E, Darden T, Prasad VR. J Biol Chem. 2006 Mar 17;281(11):7421-8. Epub 2006 Jan 18. PMID: 16423828
- Why do HIV-1 and HIV-2 use different pathways to develop AZT resistance? Boyer PL, Sarafianos SG, Clark PK, Arnold E, Hughes SH. PLoS Pathog. 2006 Feb;2(2):e10. Epub 2006 Feb 17. PMID: 16485036
- Inhibition of bacterial RNA polymerase by streptolydigin: stabilization of a straight-bridge-helix active-center conformation. Tuske S, Sarafianos SG, Wang X, Hudson B, Sineva E, Mukhopadhyay J, Birktoft JJ, Leroy O, Ismail S, Clark AD Jr, Dharia C, Napoli A, Laptenko O, Lee J, Borukhov S, Ebright RH, Arnold E. Cell. 2005 Aug 26;122(4):541-52. PMID: 16122422
- Expression and purification of SARS coronavirus proteins using SUMO-fusions. Zuo X, Mattern MR, Tan R, Li S, Hall J, Sterner DE, Shoo J, Tran H, Lim P, Sarafianos SG, Kazi L, Navas-Martin S, Weiss SR, Butt TR. Protein Expr Purif. 2005 Jul;42(1):100-10. Epub 2005 Feb 23. PMID: 15939295
- Identification of amino acid residues in the human immunodeficiency virus type-1 reverse transcriptase tryptophan-repeat motif that are required for subunit interaction using infectious virions. Mulky A, Sarafianos SG, Jia Y, Arnold E, Kappes JC. J Mol Biol. 2005 Jun 17;349(4):673-84. Epub 2005 Apr 7. PMID: 15893326
- Expression, purification, and characterization of SARS coronavirus RNA polymerase. Cheng A, Zhang W, Xie Y, Jiang W, Arnold E, Sarafianos SG, Ding J. Virology. 2005 May 10;335(2):165-76. PMID: 15840516
- Taking aim at a moving target: designing drugs to inhibit drug-resistant HIV-1 reverse transcriptases. Sarafianos SG, Das K, Hughes SH, Arnold E. Curr Opin Struct Biol. 2004 Dec;14(6):716-30. Review. PMID: 15582396
- Effects of mutations in the G tract of the human immunodeficiency virus type 1 polypurine tract on virus replication and RNase H cleavage. Julias JG, McWilliams MJ, Sarafianos SG, Alvord WG, Arnold E, Hughes SH. J Virol. 2004 Dec;78(23):13315-24. PMID: 15542682.
- Designing anti-AIDS drugs targeting the major mechanism of HIV-1 RT resistance to nucleoside analog drugs. Sarafianos SG, Hughes SH, Arnold E. Int J Biochem Cell Biol. 2004 Sep;36(9):1706-15. Review. PMID: 15183339
- Effects of the Delta67 complex of mutations in human immunodeficiency virus type 1 reverse transcriptase on nucleoside analog excision. Boyer PL, Imamichi T, Sarafianos SG, Arnold E, Hughes SH. J Virol. 2004 Sep;78(18):9987-97. PMID: 15331732
- Subunit-specific analysis of the human immunodeficiency virus type 1 reverse transcriptase in vivo. Mulky A, Sarafianos SG, Arnold E, Wu X, Kappes JC. J Virol. 2004 Jul;78(13):7089-96. PMID: 15194785
- Structures of HIV-1 RT-DNA complexes before and after incorporation of the anti-AIDS drug tenofovir. Tuske S, Sarafianos SG, Clark AD Jr, Ding J, Naeger LK, White KL, Miller MD, Gibbs CS, Boyer PL, Clark P, Wang G, Gaffney BL, Jones RA, Jerina DM, Hughes SH, Arnold E. Nat Struct Mol Biol. 2004 May;11(5):469-74. Epub 2004 Apr 25. PMID: 15107837
- Molecular model of SARS coronavirus polymerase: implications for biochemical functions and drug design. Xu X, Liu Y, Weiss S, Arnold E, Sarafianos SG, Ding J. Nucleic Acids Res. 2003 Dec 15;31(24):7117-30. PMID: 14654687
Sarafianos SG, Clark AD Jr, Das K, Tuske S, Birktoft JJ, Ilankumaran P, Ramesha AR, Sayer JM, Jerina DM, Boyer PL, Hughes SH, Arnold E. Structures of HIV-1 reverse transcriptase with pre- and post-translocation AZTMP-terminated DNA. EMBO J. 2002 Dec 2;21(23):6614-24.
Julias JG, McWilliams MJ, Sarafianos SG, Arnold E, Hughes SH. Mutations in the RNase H domain of HIV-1 reverse transcriptase affect the initiation of DNA synthesis and the specificity of RNase H cleavage in vivo. Proc Natl Acad Sci U S A. 2002 Jul 9;99(14):9515-20. Epub 2002 Jul 1.
- Tachedjian G, Orlova M, Sarafianos SG, Arnold E, Goff SP. Nonnucleoside reverse transcriptase inhibitors are chemical enhancers of dimerization of the HIV type 1 reverse transcriptase. Proc Natl Acad Sci U S A. 2001 Jun 19;98(13):7188-93.
- Boyer PL, Sarafianos SG, Arnold E, Hughes SH. Selective excision of AZTMP by drug-resistant human immunodeficiency virus reverse transcriptase. J Virol. 2001 May;75(10):4832-42.
- Sarafianos SG, Das K, Tantillo C, Clark AD Jr, Ding J, Whitcomb JM, Boyer PL, Hughes SH, Arnold E. Crystal structure of HIV-1 reverse transcriptase in complex with a polypurine tract RNA:DNA. EMBO J. 2001 Mar 15;20(6):1449-61.
- Boyer PL, Sarafianos SG, Arnold E, Hughes SH. Analysis of mutations at positions 115 and 116 in the dNTP binding site of HIV-1 reverse transcriptase. Proc Natl Acad Sci U S A. 2000 Mar 28;97(7):3056-61.
- Similarities and differences in the RNase H activities of human immunodeficiency virus type 1 reverse transcriptase and Moloney murine leukemia virus reverse transcriptase. Gao HQ, Sarafianos SG, Arnold E, Hughes SH. J Mol Biol. 1999 Dec 17;294(5):1097-113. PMID: 10600369
- Lamivudine (3TC) resistance in HIV-1 reverse transcriptase involves steric hindrance with beta-branched amino acids. Sarafianos SG, Das K, Clark AD Jr, Ding J, Boyer PL, Hughes SH, Arnold E. Proc Natl Acad Sci U S A. 1999 Aug 31;96(18):10027-32. PMID: 10468556
- Touching the heart of HIV-1 drug resistance: the fingers close down on the dNTP at the polymerase active site. Sarafianos SG, Das K, Ding J, Boyer PL, Hughes SH, Arnold E. Chem Biol. 1999 May;6(5):R137-46. Review. PMID: 10322129
- Structure and functional implications of the polymerase active site region in a complex of HIV-1 RT with a double-stranded DNA template-primer and an antibody Fab fragment at 2.8 A resolution. Ding J, Das K, Hsiou Y, Sarafianos SG, Clark AD Jr, Jacobo-Molina A, Tantillo C, Hughes SH, Arnold E. J Mol Biol. 1998 Dec 11;284(4):1095-111. PMID: 9837729
- Role of glutamine-151 of human immunodeficiency virus type-1 reverse transcriptase in RNA-directed DNA synthesis. Kaushik N, Harris D, Rege N, Modak MJ, Yadav PN, Pandey VN. Biochemistry. 1997 Nov 25;36(47):14430-8. PMID: 9398161
- Mechanism of polyoxometalate-mediated inactivation of DNA polymerases: an analysis with HIV-1 reverse transcriptase indicates specificity for the DNA-binding cleft. Sarafianos SG, Kortz U, Pope MT, Modak MJ. Biochem J. 1996 Oct 15;319 ( Pt 2):619-26. PMID: 8912703
- Biochemical analysis of catalytically crucial aspartate mutants of human immunodeficiency virus type 1 reverse transcriptase. Kaushik N, Rege N, Yadav PN, Sarafianos SG, Modak MJ, Pandey VN. Biochemistry 1996 Sep 10;35(36):11536-46. PMID: 8794733
- Role of methionine 184 of human immunodeficiency virus type-1 reverse transcriptase in the polymerase function and fidelity of DNA synthesis. Pandey VN, Kaushik N, Rege N, Sarafianos SG, Yadav PN, Modak MJ. Biochemistry 1996 Feb 20;35(7):2168-79. PMID: 8652558
- Site-directed mutagenesis of arginine 72 of HIV-1 reverse transcriptase. Catalytic role and inhibitor sensitivity. Sarafianos SG, Pandey VN, Kaushik N, Modak MJ. J Biol Chem. 1995 Aug 25;270(34):19729-35. PMID: 7544345
- Glutamine 151 participates in the substrate dNTP binding function of HIV-1 reverse transcriptase. Sarafianos SG, Pandey VN, Kaushik N, Modak MJ. Biochemistry. 1995 May 30;34(21):7207-16. PMID: 7539293
AT32P-dependent estimation of nanomoles of fatty acids: its use in the assay of phospholipase A2 activity. Sarafianos SG, Nair PP, Kumar S. Anal Biochem. 1990 May 1;186(2):374-9. PMID: 2163589